In 2024, we’ll have much to look at in our wonderful solar system.


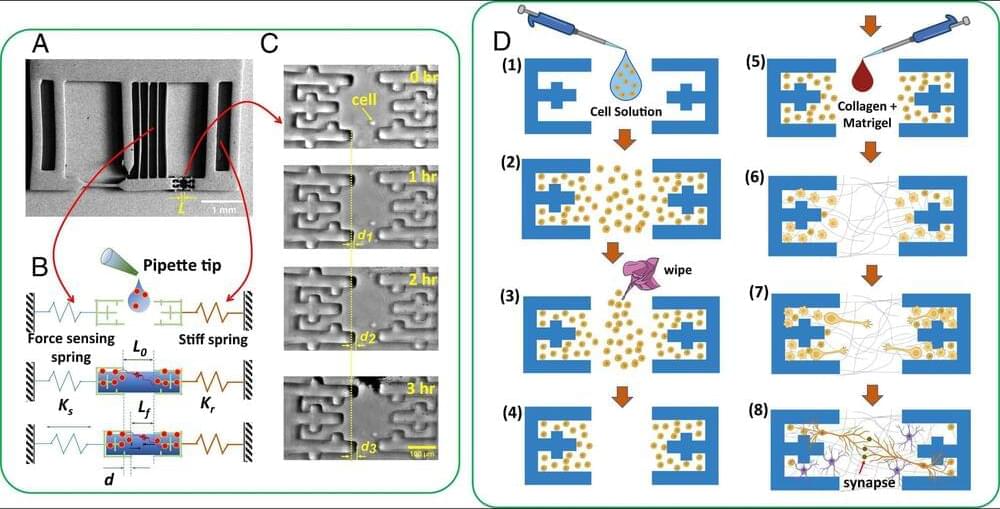
Neurons in the brain communicate with each other at their synapses. It has long been understood that this communication occurs through biochemical processes. Here, we reveal that mechanical tension in neurons is essential for communication. Using in vitro rat hippocampal neurons, we find that 1) neurons become tout/tensed after forming synapses resulting in a contractile neural network, and 2) without this contractility, neurons fail to fire. To measure time evolution of network contractility in 3D (not 2D) extracellular matrix, we developed an ultrasensitive force sensor with 1 nN resolution. We employed Multi-Electrode Array and iGluSnFR, a glutamate sensor, to quantify neuronal firing at the network and at the single synapse scale, respectively. When neuron contractility is relaxed, both techniques show significantly reduced firing. Firing resumes when contractility is restored.

This year gave rise to an incredible mix of brain implants that can record, decode, and alter brain activity.
It sounds like déjà vu—brain-machine interfaces also lived rent free in my head in last year’s roundup, but for good reason. Neuroscientists are building increasingly sophisticated and flexible electronic chips that seamlessly integrate machine intelligence with our brains and spinal cords at record-breaking speed. What was previously science fiction—for example, helping paralyzed people regain their ability to walk, swim, and kayak—is now reality.
This year, brain implants further transformed people’s lives. The not-so-secret sauce? AI.
Google is reportedly planning to cut 30,000 jobs by integrating artificial intelligence into daily operations. 2023 has witnessed mass layoffs triggered by the emergence of AI replacements. Palki Sharma explains how you can save your job from an AI takeover. — Google | Mass Layoffs | Artificial Intelligence | Job Opportunities | Firstpost | World News | Vantage | Palki Sharma #google #layoffs #jobmarket #ai #artificialintelligence #technology #firstpost #vantageonfirstpost #palkisharma #worldnews Vantage is a ground-breaking news, opinions, and current affairs show from Firstpost. Catering to a global audience, Vantage covers the biggest news stories from a 360-degree perspective, giving viewers a chance to assess the impact of world events through a uniquely Indian lens. The show is anchored by Palki Sharma, Managing Editor, Firstpost. By breaking stereotypes, Vantage aims to challenge conventional wisdom and present an alternative view on global affairs, defying the norm and opening the door to new perspectives. The show goes beyond the headlines to uncover the hidden stories – making Vantage a destination for thought-provoking ideas. Vantage airs Monday to Friday at 9 PM IST on Firstpost across all leading platforms. Subscribe to Firstpost channel and press the bell icon to get notified when we go live. / @firstpost Follow Firstpost on Instagram:
/ firstpost Follow Firstpost on Facebook:
/ firstpostin Follow Firstpost on Twitter:
/ firstpost Follow Firstpost on WhatsApp: https://www.whatsapp.com/channel/0029…
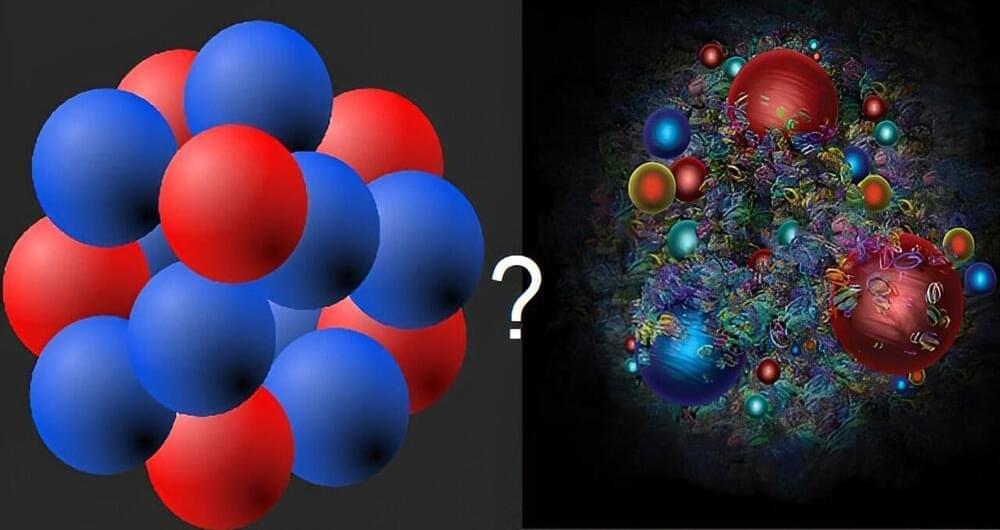
Atomic nuclei are made of nucleons (like protons and neutrons), which themselves are made of quarks. When crushed at high densities, nuclei dissolve into a liquid of nucleons and, at even higher densities, the nucleons themselves dissolve into a quark liquid.
In a new study, published in the journal Physical Review B, researchers addressed the question of whether the liquids of nucleons and quarks are fundamentally different.
Their theoretical calculations suggest that these liquids are different. Both types of liquids produce vortices when they rotate, but in quark liquids, the vortices carry a “color-magnetic field,” similar to an ordinary magnetic field. There is no such effect in nucleon liquids. Thus, these vortices sharply distinguish quark liquids from nuclear liquids.
Alter 3 has just been unveiled by the University of Tokyo and its powered by GPT-4, capable of human-like activities and interpreting verbal instructions. Researchers at the Technical University of Munich developed a self-aware robot with proprioception, enhancing its movement and interaction capabilities. The University of Southern California introduced RoboCLIP, an algorithm that trains robots to perform tasks in new environments with minimal instruction. Intel Labs and partners created advanced motor control for robots using hierarchical generative models, significantly improving their ability to perform complex tasks.\
\
Deep Learning AI Specialization: https://imp.i384100.net/GET-STARTED\
AI Marketplace: https://taimine.com/\
\
AI news timestamps:\
0:00 Alter 3 GPT4 powered AI robot\
1:31 Robot self awareness\
3:30 RoboCLIP\
5:22 Motor control for autonomous robots\
\
#ai #robot #technology
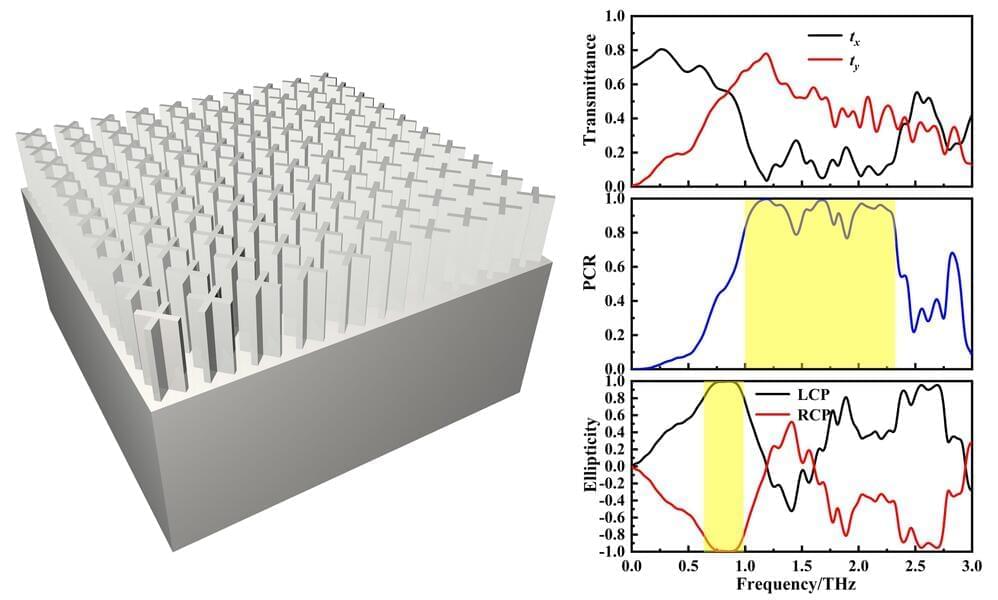
Polarization is one of the fundamental characteristics of electromagnetic waves. It can convey valuable vector information in sensitive measurements and signal transmission, which is a promising technology for various fields such as environmental monitoring, biomedical sciences, and marine exploration. Particularly in the terahertz frequency range, traditional device design methods and structures can only achieve limited performance. Designing efficient modulator devices for high-bandwidth terahertz waves presents a significant challenge.
Researchers led by Prof. Liang Wu at Tianjin University (TJU), China, have been conducting experiments in the field of all-dielectric metamaterials, specifically focusing on utilizing these materials and their structural design to achieve effective broadband polarization conversion in the terahertz frequency range.
They propose a cross-shaped microstructure metamaterial for achieving cross-polarization conversion and linear-to-circular polarization conversion in the terahertz frequency range. The study, titled “An all-silicon design of a high-efficiency broadband transmissive terahertz polarization convertor,” was published in Frontiers of Optoelectronics.
