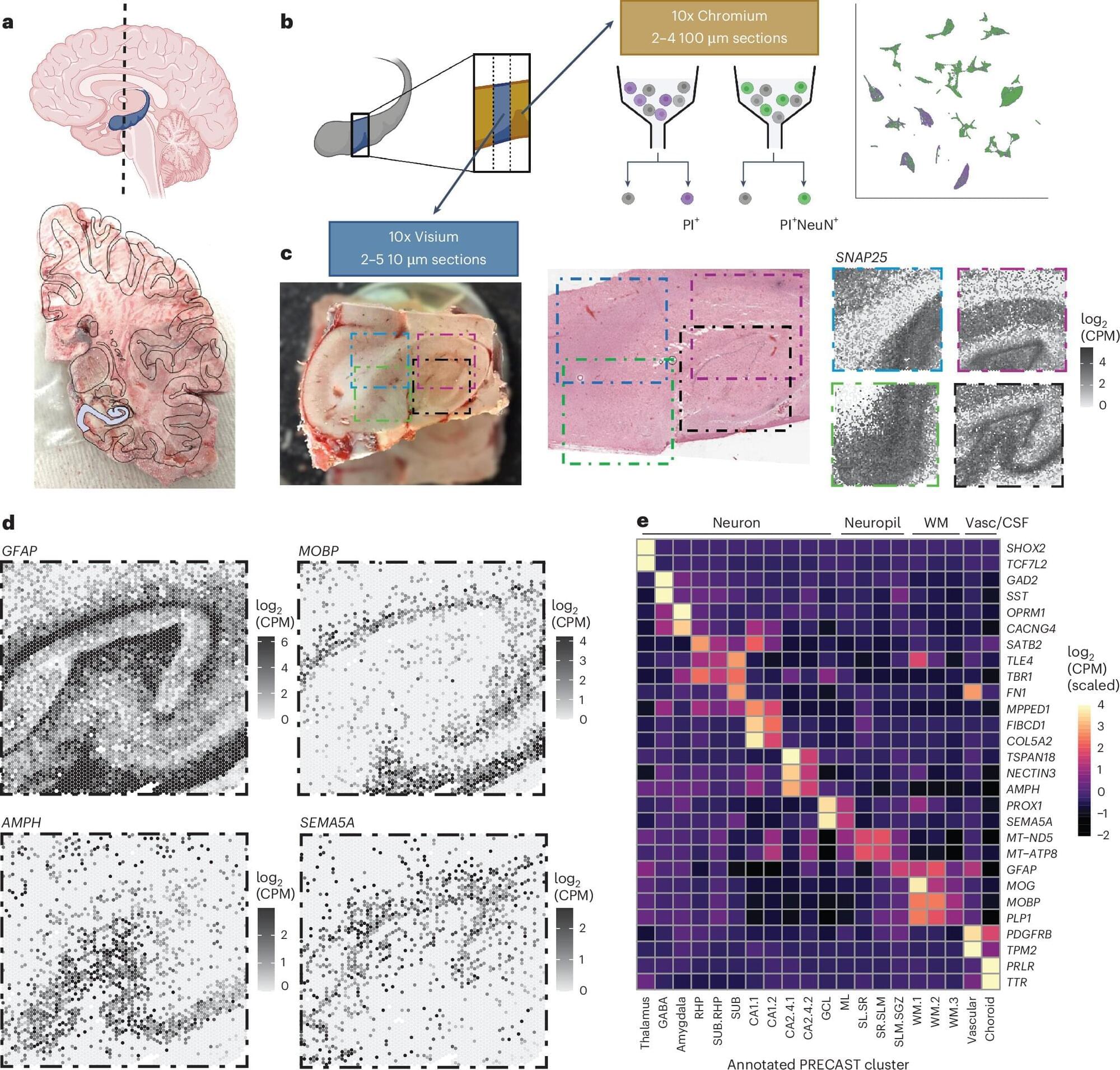
The hippocampus is an important brain region known to support various cognitive (i.e., mental) processes, including the encoding and retrieval of memories, learning, decision-making and the regulation of emotional states. While extensive research has tried to delineate the structure, functions and organization of the hippocampus, the cell types contained within it and their connections with other neurons have not yet been fully mapped out.
Over the past decades, available methods for studying cell subpopulations, the expressions of genes within them and their connectivity have become increasingly advanced. One of these methods, known as spatially resolved transcriptomics, works by measuring the expression of genes in cells while preserving their arrangement in space. Another called single-nucleus RNA-sequencing (snRNA-seq), allows scientists to examine RNA molecules inside individual cell nuclei to detect differences between them and categorize cells into different subtypes.
Researchers at Johns Hopkins Bloomberg School of Public Health, the Lieber Institute for Brain Development and Johns Hopkins School of Medicine recently used a combination of these two experimental techniques to examine cells in tissue extracted from the hippocampus. Their paper, published in Nature Neuroscience, introduces a comprehensive molecular atlas of the hippocampus that maps different cell subtypes and their organization.