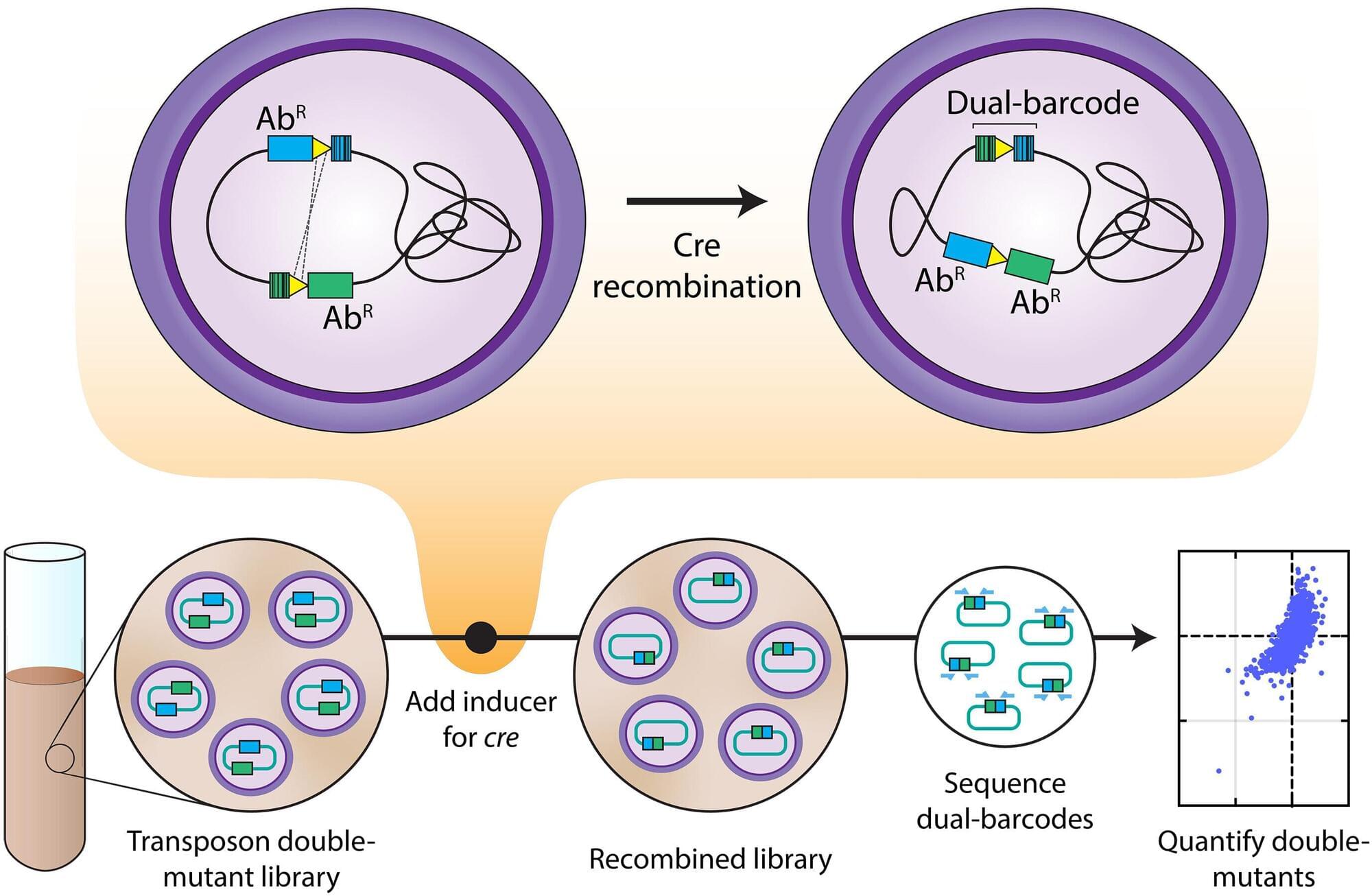
Despite rapid advances in reading the genetic code of living organisms, scientists still face a major challenge today—knowing a gene’s sequence does not automatically reveal what it does. Even in simple, well-studied bacteria like Escherichia coli (better known as E. coli), about one-quarter of the genes have no known function. Traditional approaches—turning off one gene at a time and studying the effects—are slow, laborious, and sometimes inconclusive due to gene redundancy.
Researchers from the Yong Loo Lin School of Medicine, National University of Singapore (NUS Medicine) and collaborators from the University of California, Berkeley (UC Berkeley) have developed a new technique called Dual transposon sequencing (Dual Tn-seq), which allows for rapid identification of genetic interactions. It maps how bacterial genes work together, revealing vulnerabilities that could be targeted by future antibiotics.
“This is like mapping the social network for bacterial genes,” said Assistant Professor Chris Sham Lok To from the Infectious Diseases Translational Research Program and the Department of Microbiology and Immunology, NUS Medicine, who led the study. “We can now see which genes depend on each other, and which pairs of genes bacteria can’t live without. That’s exactly the insight we need for next-generation antibiotics.”