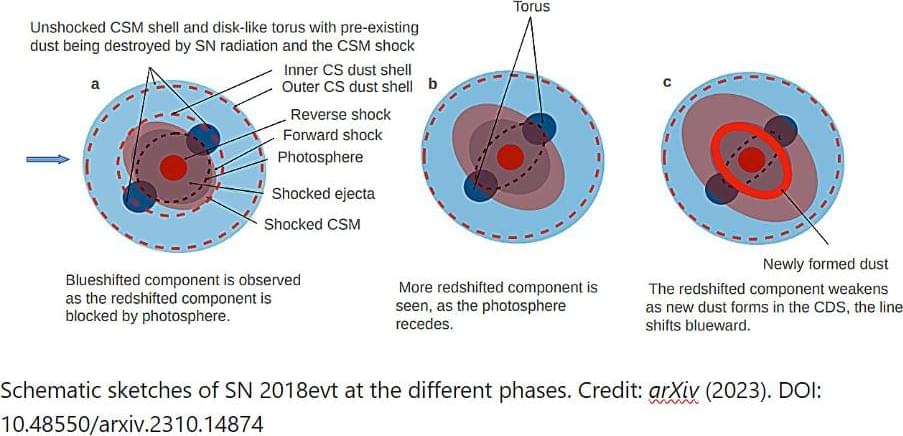
Cosmic dust—like dust on Earth—comprises groupings of molecules that have condensed and stuck together in a grain. But the exact nature of dust creation in the universe has long been a mystery. Now, however, an international team of astronomers from China, the United States, Chile, the United Kingdom, Spain, etc., has made a significant discovery by identifying a previously unknown source of dust in the universe: a Type 1a supernova interacting with gas from its surroundings.
The study was published in Nature Astronomy on Feb. 9, and was led by Prof. Wang Lingzhi from the South America Center for Astronomy of the Chinese Academy of Sciences.
Supernovae have been known to play a role in dust formation, and to date, dust formation has only been seen in core-collapse supernovae—the explosion of massive stars. Since core-collapse supernovae do not occur in elliptical galaxies, the nature of dust creation in such galaxies has remained elusive.