Book by Nick Bostrom: Chapter Summary, Free PDF Download, Audiobook, Review. Charting the Future of AI: Risks and Imperatives.




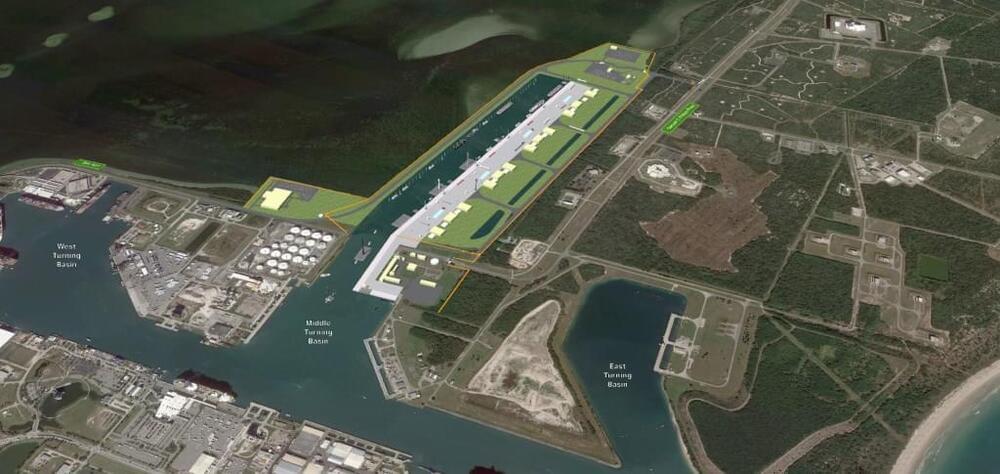
More Space Coast rocket launches mean a crowded fleet of support ships are already pushing Port Canaveral’s limits. So the state commissioned a study that suggests a $2.1 billion solution to give companies such as SpaceX and Blue Origin dedicated places to dock.
Space Florida, the state’s aerospace finance and development authority, released Thursday the Florida Spaceport System Maritime Intermodal Transportation Study that’s been in the works for more than a year.
“It was really important for us to make sure we had every stakeholder input throughout the process,” said Space Florida president and CEO Rob Long.

Daniel Dennett, who died in April at the age of 82, was a towering figure in the philosophy of mind. Known for his staunch physicalist stance, he argued that minds, like bodies, are the product of evolution. He believed that we are, in a sense, machines—but astoundingly complex ones, the result of millions of years of natural selection.
Dennett wrote more than a dozen books, some of them aimed at a scholarly audience but many of them directed squarely at the inquisitive non-specialist—including bestsellers like Consciousness Explained, Breaking the Spell, and Darwin’s Dangerous Idea. Reading his works, one gets the impression of a mind jammed to the rafters with ideas. As Richard Dawkins put it in a blurb for Dennett’s last book, a memoir titled I’ve Been Thinking: “How unfair for one man to be blessed with such a torrent of stimulating thoughts.”

A professor at the University of Warwick is exploring the chemistry of the galaxy far, far away this Star Wars Day, May the 4th.
Science fiction is meeting science fact, as Professor Alex Baker discusses the captivating inspiration real-world reactions have had on the Star Wars universe.
The chemist from the University of Warwick explores what may underpin the freezing of Han Solo, the colors of lightsabers, the reactions that power star ships and much more.
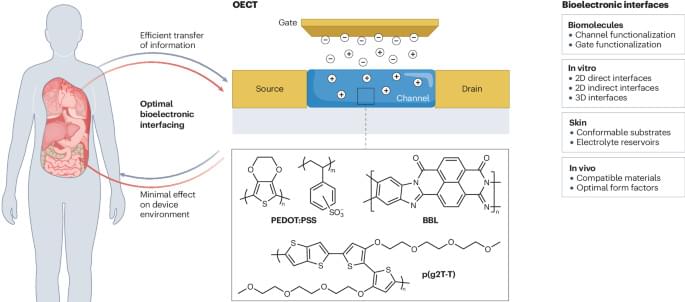
The organic electrochemical transistor stands out as a tool for constructing powerful biosensors owing to its high signal transduction ability and adaptability to various geometrical forms. However, the performance of organic electrochemical transistors relies on stable and seamless interfaces with biological systems. This Review examines strategies to improve and optimize interfaces between organic electrochemical transistors and various biological components.
Researchers have successfully used 40-year-old mathematics to explain quantum tunneling, providing a unified approach to diverse quantum phenomena.
Quantum mechanical effects such as radioactive decay, or more generally: ‘tunneling’, display intriguing mathematical patterns. Two researchers at the University of Amsterdam now show that a 40-year-old mathematical discovery can be used to fully encode and understand this structure.
Quantum Physics – Easy and Hard.
Scientists have only begun to discover the endless possibilities hidden within our universe, like finding an entire galaxy that shouldn’t exist! Join us in today’s epic new video as we explore an impossible galaxy!
🔔 SUBSCRIBE TO THE INFOGRAPHICS SHOW ►
🔖 MY SOCIAL PAGES
TikTok ► / theinfographicsshow.
Discord ► / discord.
Facebook ► / theinfographicsshow.
Twitter ► / theinfoshow.
💭 Find more interesting stuff on:
https://www.theinfographicsshow.com.
📝 SOURCES: https://pastebin.com/ribHP3qp.
All videos are based on publicly available information unless otherwise noted.