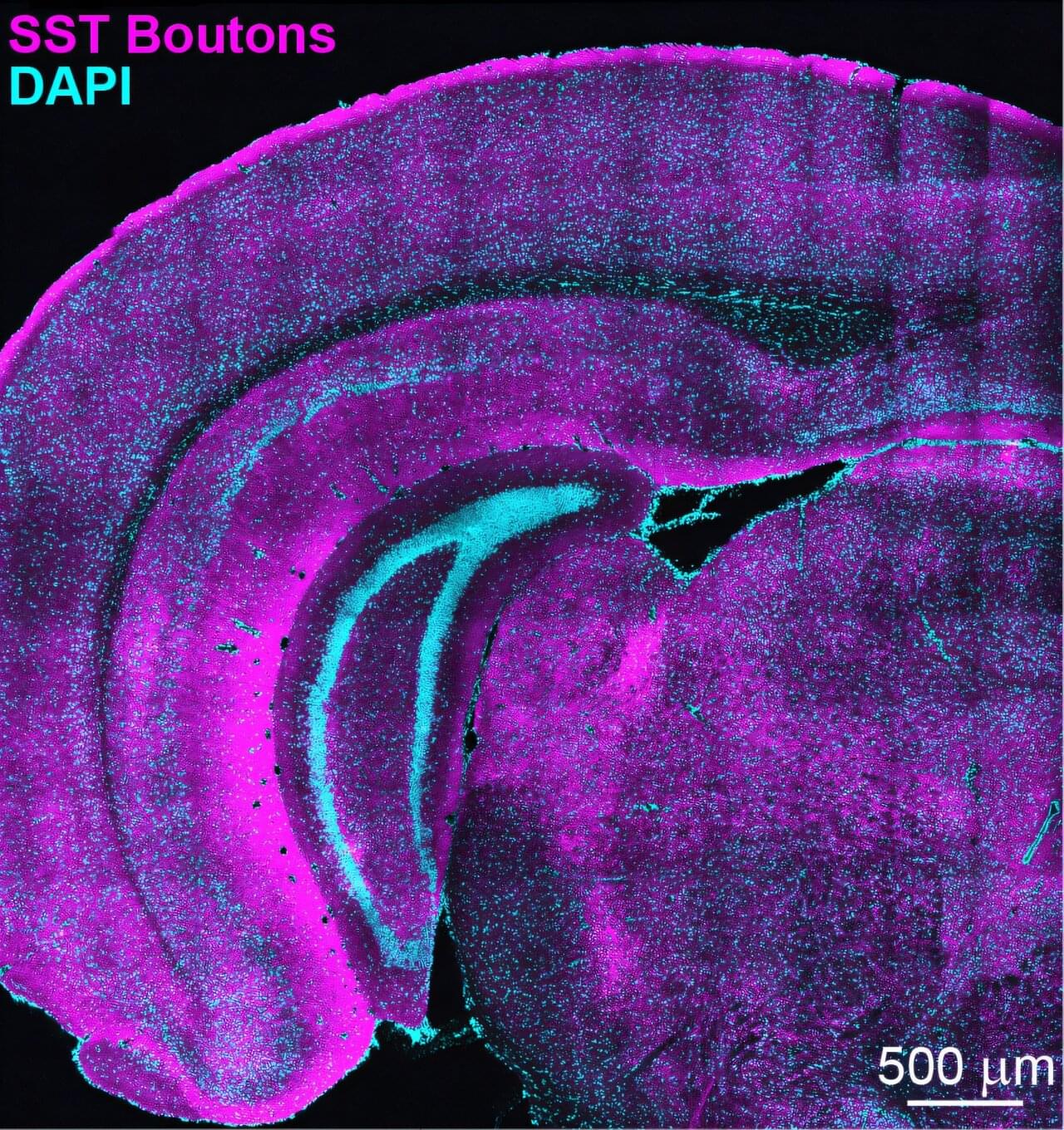
The way the brain develops can shape us throughout our lives, so neuroscientists are intensely curious about how it happens. A new study by researchers in The Picower Institute for Learning and Memory at MIT that focused on visual cortex development in mice, reveals that an important class of neurons follows a set of rules that while surprising, might just create the right conditions for circuit optimization.
During early brain development, multiple types of neurons emerge in the visual cortex (where the brain processes vision). Many are “excitatory,” driving the activity of brain circuits, and others are “inhibitory,” meaning they control that activity. Just like a car needs not only an engine and a gas pedal, but also a steering wheel and brakes, a healthy balance between excitation and inhibition is required for proper brain function.
During a “critical period” of development in the visual cortex, soon after the eyes first open, excitatory and inhibitory neurons forge and edit millions of connections, or synapses, to adapt nascent circuits to the incoming flood of visual experience. Over many days, in other words, the brain optimizes its attunement to the world.