The NASA Sun Science probe is now preparing for its next solar flyby in April 2019: https://go.nasa.gov/2FSvd5H
Get the latest international news and world events from around the world.


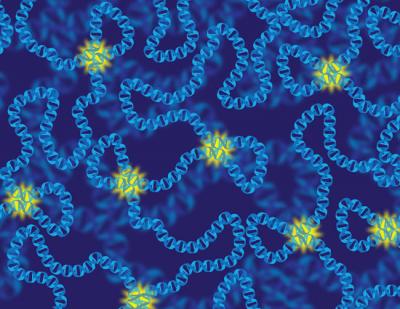

All-photonic quantum repeaters could lead to a faster, more secure global quantum internet
Engineering researchers have demonstrated proof-of-principle for a device that could serve as the backbone of a future quantum Internet. University of Toronto Engineering professor Hoi-Kwong Lo and his collaborators have developed a prototype for a key element for all-photonic quantum repeaters, a critical step in long-distance quantum communication.

The American Public Increasingly Desires Life Extension
Attitudes about life extension have significantly changed within the last decade.
While medical schools have had the idea that aging should be brought under medical control for over a century, the explicit desire to greatly extend one’s life remained rare – until very recently. A new study by YouGov, a market intelligence company that researches multiple topics, found that, today, one in five Americans agrees with the statement “I want to live forever.” Is this the result of some sort of bias, or does it mean that we are reaching a turning point, after which society will start boldly and unambiguously clamoring for the cure for aging?
The desire for a long life
The wish to remain healthy and young and live significantly longer has accompanied humanity since its dawn. The most ancient myths and the very first pieces of literature, such as the Epic of Gilgamesh, tried to explain human aging and mortality, and they pictured heroes who pursued ways to live indefinitely or to save their loved ones. These early motivations are woven into modern religions, and they eventually caused life and health to be considered universal human rights that have to be valued and protected.


In test of wisdom, new research favours Yoda over Spock
Wise reasoning does not necessarily require uniform emotional control or suppression, says Igor Grossmann, professor of psychology at Waterloo and lead author of the new study. Instead, wise reasoning can also benefit from a rich and balanced emotional life.
A person’s ability to reason wisely about a challenging situation may improve when they also experience diverse yet balanced emotions, say researchers from the University of Waterloo.
The finding clarifies millennia of philosophical and psychological thinking that debates how wisdom is related to the effective management of emotionally charged experiences.