On August 21, 2023, my 70th birthday, I, Rev. Ivan Stang, used RunwayML and Wombo Dream on my phone to make this A.I. video for the classic song \.
Category: robotics/AI – Page 305
Fake job seekers are flooding U.S. companies that are hiring for remote positions, tech CEOs say
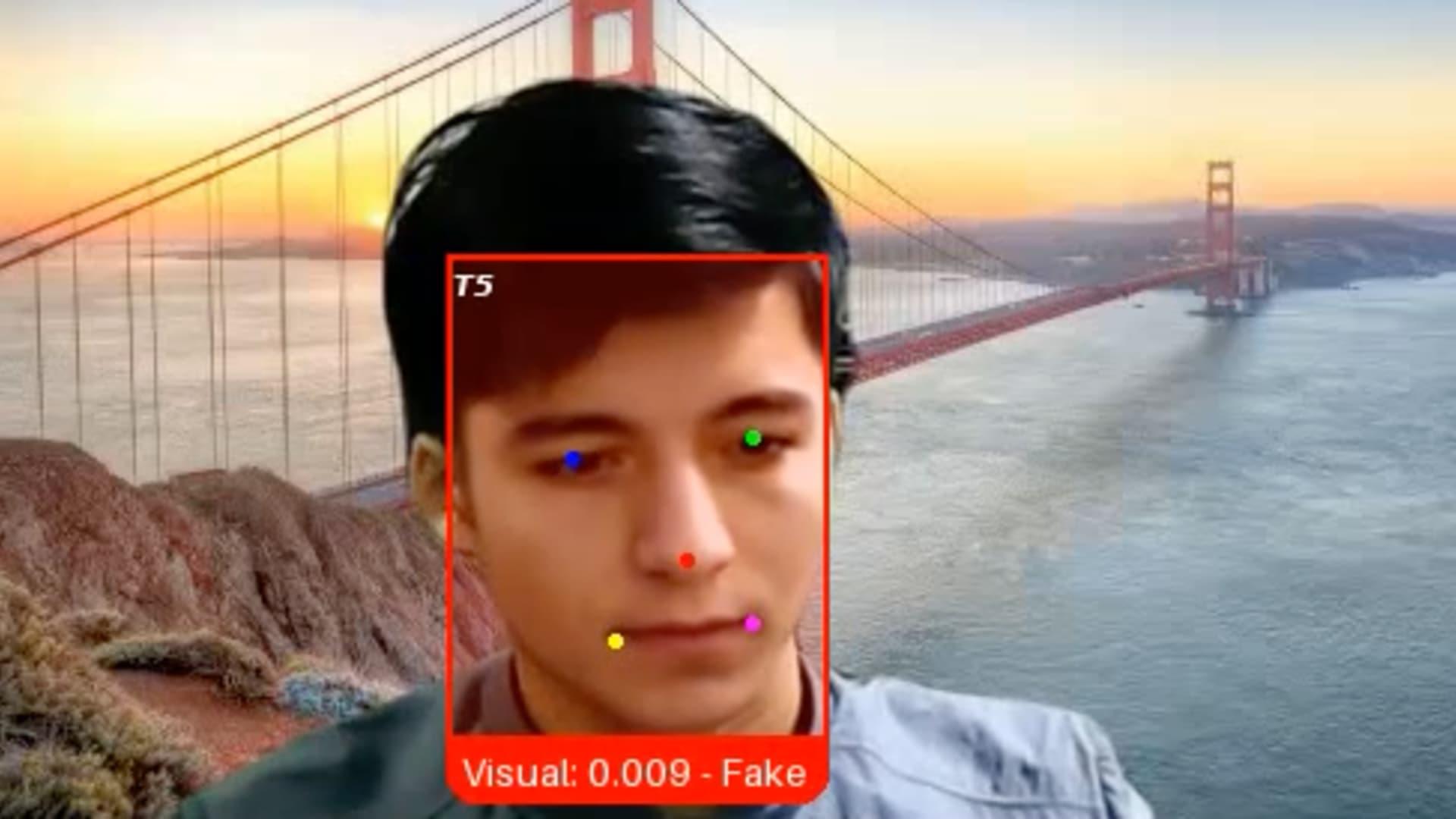
That’s because the candidate, whom the firm has since dubbed “Ivan X,” was a scammer using deepfake software and other generative AI tools in a bid to get hired by the tech company, said Pindrop CEO and co-founder Vijay Balasubramaniyan.
“Gen AI has blurred the line between what it is to be human and what it means to be machine,” Balasubramaniyan said. “What we’re seeing is that individuals are using these fake identities and fake faces and fake voices to secure employment, even sometimes going so far as doing a face swap with another individual who shows up for the job.”
Companies have long fought off attacks from hackers hoping to exploit vulnerabilities in their software, employees or vendors. Now, another threat has emerged: Job candidates who aren’t who they say they are, wielding AI tools to fabricate photo IDs, generate employment histories and provide answers during interviews.
AI for the Enterprise | Dennis Wilson, Founder DBC Technologies | Inbound & Outbound AI Voice Agents
In this second episode of the (A)bsolutely (I)ncredible Podcast, I sit down with Dennis Wilson, Founder of DBC Technologies.
Dennis is deeply involved in my friend Jim Roddy’s Retail Solution Providers Association (RSPA) and is a regular speaker at RSPA events.
Dennis shares the benefits of AI with these providers. He is a passionate marketer who has created a platform that utilizes the best Al capabilities the industry marketplace has to offer.
DBC stands for Doing Business Creatively — utilizing 25 + years of CRM, software, marketing, and sales automation experience.
DBC has been deeply involved and integrating Al into their software and client’s businesses since the launch of OpenAI’s ChatGTP.
If you’re interested in AI Voice solutions for your business let me know and I’ll be glad to connect you with Dennis and his team of experts.
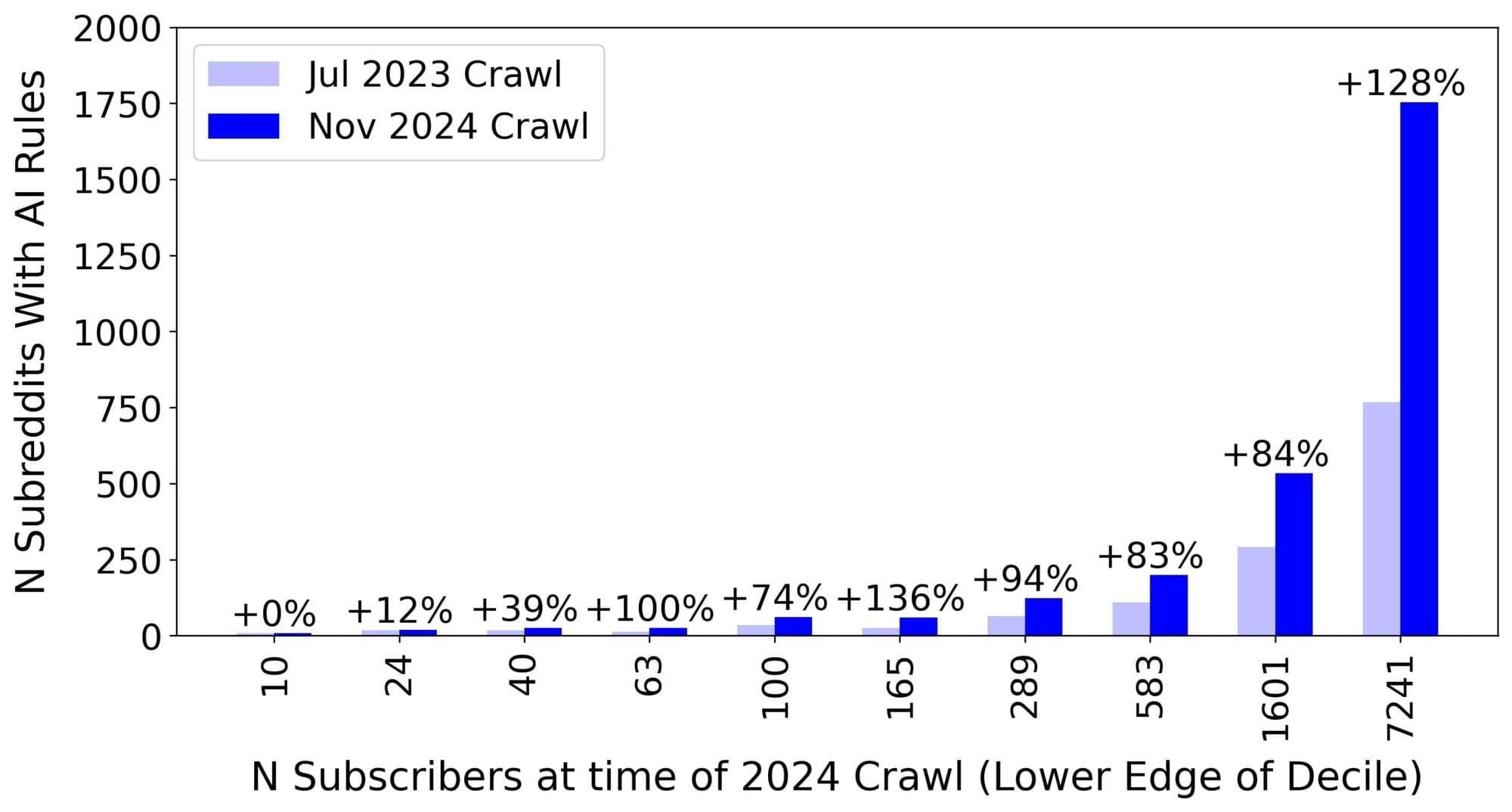
Dataset reveals how Reddit communities are adapting to AI
Researchers at Cornell Tech have released a dataset extracted from more than 300,000 public Reddit communities, and a report detailing how Reddit communities are changing their policies to address a surge in AI-generated content.
The team collected metadata and community rules from the online communities, known as subreddits, during two periods in July 2023 and November 2024. The researchers will present a paper with their findings at the Association of Computing Machinery’s CHI conference on Human Factors in Computing Systems being held April 26 to May 1 in Yokohama, Japan.
One of the researchers’ most striking discoveries is the rapid increase in subreddits with rules governing AI use. According to the research, the number of subreddits with AI rules more than doubled in 16 months, from July 2023 to November 2024.
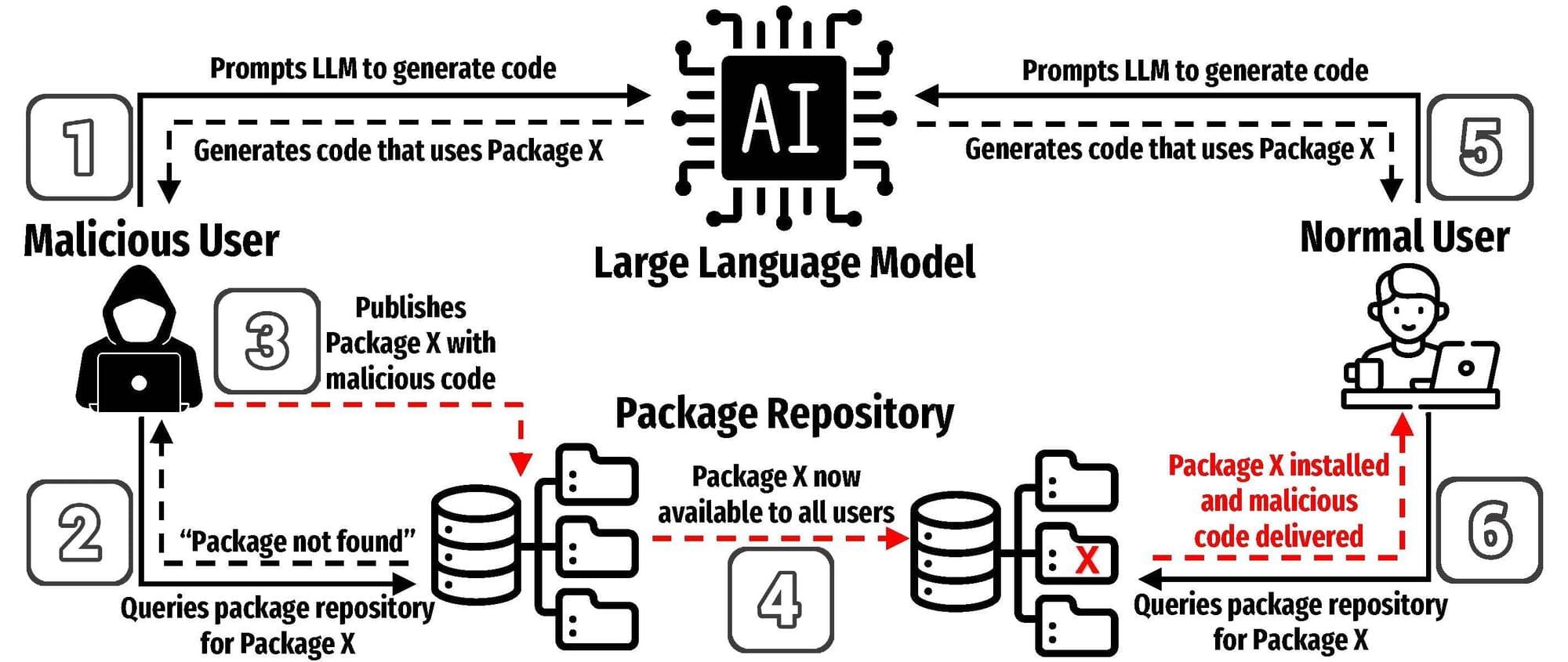
AI threats in software development revealed in new study
UTSA researchers recently completed one of the most comprehensive studies to date on the risks of using AI models to develop software. In a new paper, they demonstrate how a specific type of error could pose a serious threat to programmers that use AI to help write code.
Joe Spracklen, a UTSA doctoral student in computer science, led the study on how large language models (LLMs) frequently generate insecure code.
His team’s paper, published on the arXiv preprint server, has also been accepted for publication at the USENIX Security Symposium 2025, a cybersecurity and privacy conference.

Is AI truly creative? Study shows how visibility of process shapes perception
What makes people think an AI system is creative? New research shows that it depends on how much they see of the creative act. The findings have implications for how we research and design creative AI systems, and they also raise fundamental questions about how we perceive creativity in other people.
The work is published in the journal ACM Transactions on Human-Robot Interaction.
“AI is playing an increasingly large role in creative practice. Whether that means we should call it creative or not is a different question,” says Niki Pennanen, the study’s lead author. Pennanen is researching AI systems at Aalto University and has a background in psychology. Together with other researchers at Aalto and the University of Helsinki, he did experiments to find out whether people think a robot is more creative if they see more of the creative act.
Swiss-UK team rolls out robot dogs to transform doorstep deliveries
RIVR and Evri debut wheeled-legged delivery robots in the UK, aiming to transform last-mile logistics with smart, scalable automation.


Amazon says its AI video model can now generate minutes-long clips
Amazon has upgraded its AI video model, Nova Reel, with the ability to generate videos up to two minutes in length.
Nova Reel, announced in December 2024, was Amazon’s first foray into the generative video space. It competes with models from OpenAI, Google, and others in what’s fast becoming a crowded market.
The latest Nova Reel, Nova Reel 1.1, can generate “multi-shot” videos with “consistent style” across shots, explained AWS developer advocate Elizabeth Fuentes in a blog post. Users can provide a prompt up to 4,000 characters long to generate up to a two-minute video composed of six-second shots.
New AI Tool, MindGlide, Uses Existing MRI Scans to Unlock MS Insights
The results from the study show that it is possible to use MindGlide to accurately identify and measure important brain tissues and lesions even with limited MRI data and single types of scans that aren’t usually used for this purpose—such as T2-weighted MRI without FLAIR (a type of scan that highlights fluids in the body but still contains bright signals, making it harder to see plaques). “Our results demonstrate that clinically meaningful tissue segmentation and lesion quantification are achievable even with limited MRI data and single contrasts not typically used for these tasks (e.g., T2-weighted MRI without FLAIR),” they stated. “Importantly, our findings generalized across datasets and MRI contrasts. Our training used only FLAIR and T1 images, yet the model successfully processed new contrasts (like PD and T2) from different scanners and periods encountered during external validation.”
As well as performing better at detecting changes in the brain’s outer layer, MindGlide also performed well in deeper brain areas. The findings were valid and reliable both at one point in time and over longer periods (i.e., at annual scans attended by patients). Additionally, MindGlide was able to corroborate previous high-quality research regarding which treatments were most effective. “In clinical trials, MindGlide detected treatment effects on T2-lesion accrual and cortical and deep grey matter volume loss. In routine-care data, T2-lesion volume increased with moderate-efficacy treatment but remained stable with high-efficacy treatment,” the investigators wrote in summary.
The researchers now hope that MindGlide can be used to evaluate MS treatments in real-world settings, overcoming previous limitations of relying solely on high-quality clinical trial data, which often did not capture the full diversity of people with MS.