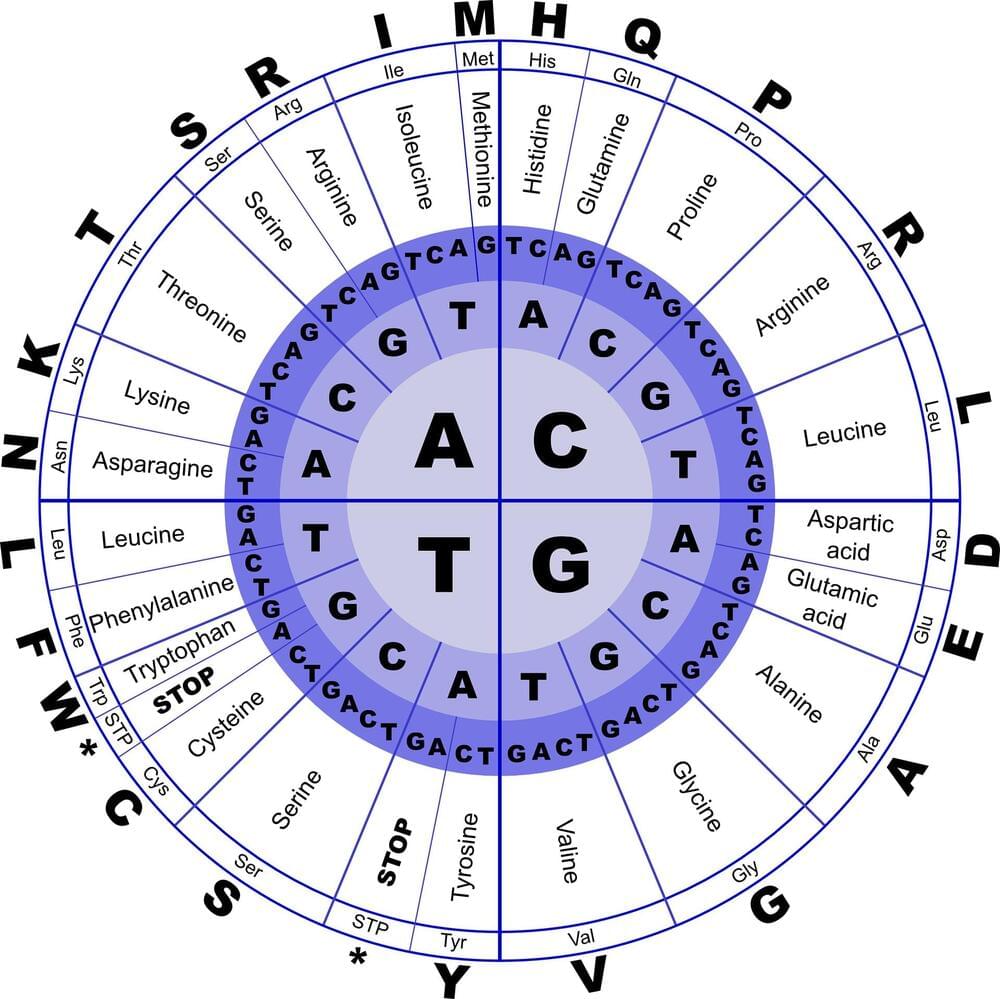
Basic biology textbooks will tell you that all life on Earth is built from four types of molecules: proteins, carbohydrates, lipids, and nucleic acids. And each group is vital for every living organism.
But what if humans could actually show that these “molecules of life,” such as amino acids and DNA bases, can be formed naturally in the right environment? Researchers at the University of Florida are using the HiPerGator—the fastest supercomputer in U.S. higher education—to test this experiment.
HiPerGator—with its AI models and vast capacity for graphics processing units, or GPUs (specialized processors designed to accelerate graphics renderings)—is transforming the molecular research game.