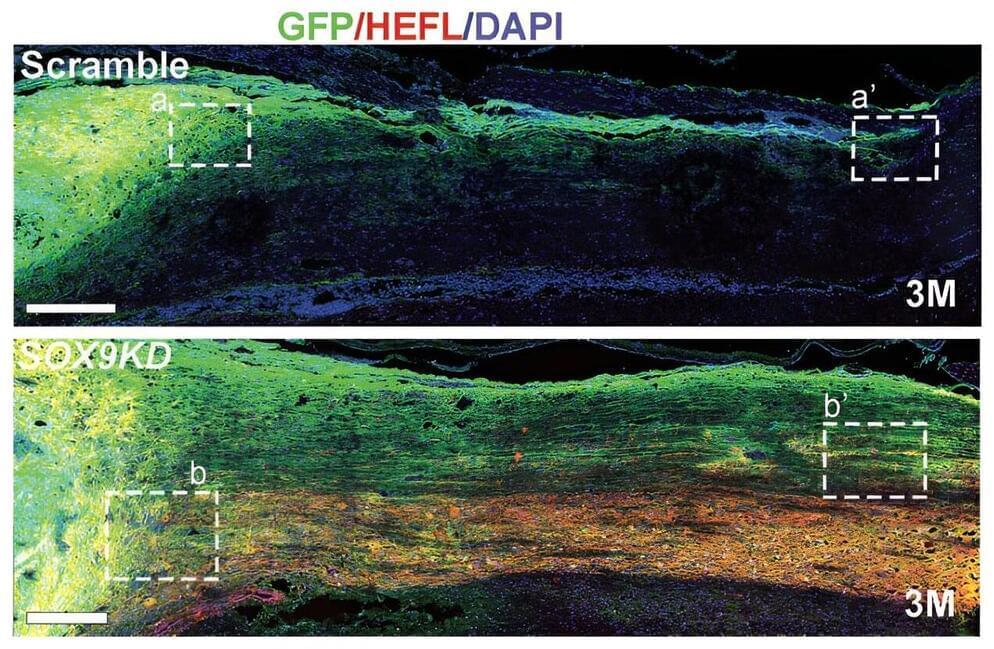
A research team co-led by City University of Hong Kong (CityU) and The University of Hong Kong (HKU) has recently made a significant advancement in spinal cord injury treatment by using genetically-modified human neural stem cells (hNSCs).
They found that specifically modulating a gene expression to a certain level in hNSCs can effectively promote the reconstruction of damaged neural circuits and restore locomotor functions, offering great potential for new therapeutic opportunities for patients with spinal cord injury. The findings were published in the journal Advanced Science under the title “Transplanting Human Neural Stem Cells with ≈50% Reduction of SOX9 Gene Dosage Promotes Tissue Repair and Functional Recovery from Severe Spinal Cord Injury.”
Traumatic spinal cord injury is a devastating condition that commonly results from accidents such as falls, car crashes or sport-related injuries.