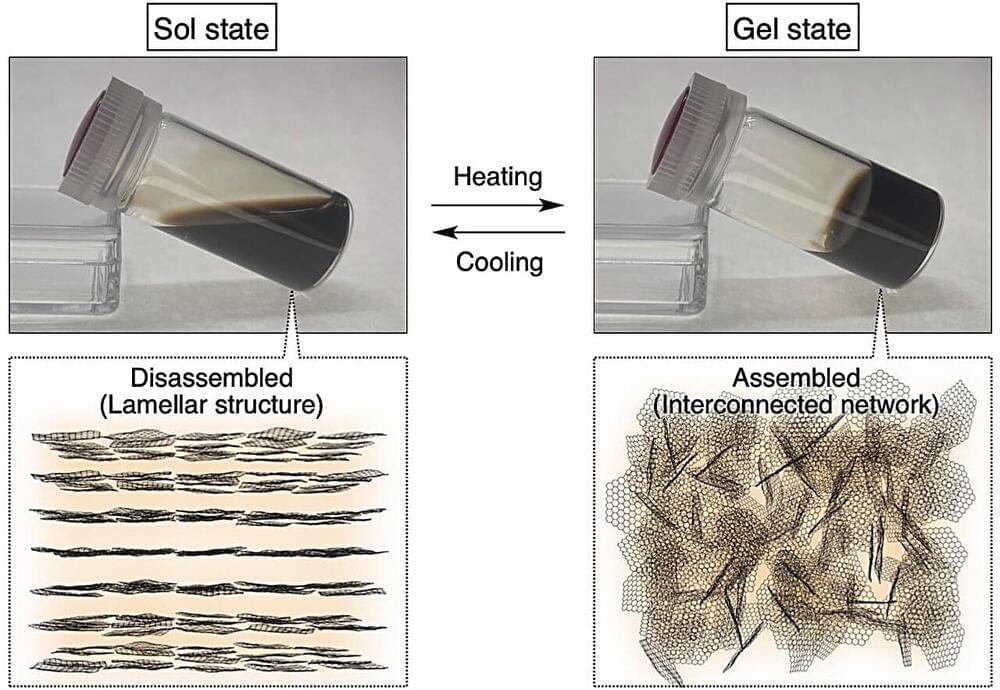
Graphene-based two-dimensional materials have recently emerged as a focus of scientific exploration due to their exceptional structural, mechanical, electrical, optical, and thermal properties. Among them, nanosheets based on graphene-oxide (GO), an oxidized derivative of graphene, with ultrathin and extra wide dimensions and oxygen-rich surfaces are quite promising.
Functional groups containing oxygen, such as carboxy and acidic hydroxy groups, generate dense negative charges, making GO nanosheets colloidally stable in water. As a result, they are valuable building blocks for next-generation functional soft materials.
In particular, thermoresponsive GO nanosheets have garnered much attention for their wide-ranging applications, from smart membranes and surfaces and recyclable systems to hydrogel actuators and biomedical platforms. However, the prevailing synthetic strategies for generating thermoresponsive behaviors entail modifying GO nanosheet surfaces with thermoresponsive polymers such as poly (N-isopropylacrylamide). This process is complex and has potential limitations in subsequent functionalization efforts.