Researchers at the First Affiliated Hospital of Chongqing Medical University in China have uncovered a sharply rising burden of skin cancer in older adults driven largely by population growth and affecting men twice as often.
Skin cancer already ranks among the costliest malignancies to treat, and an aging world means more time for ultraviolet damage to accumulate. Previous research shows older patients now make up nearly three-quarters of new cases, yet global data capturing the full scope and trend in those over 65 remains scarce.
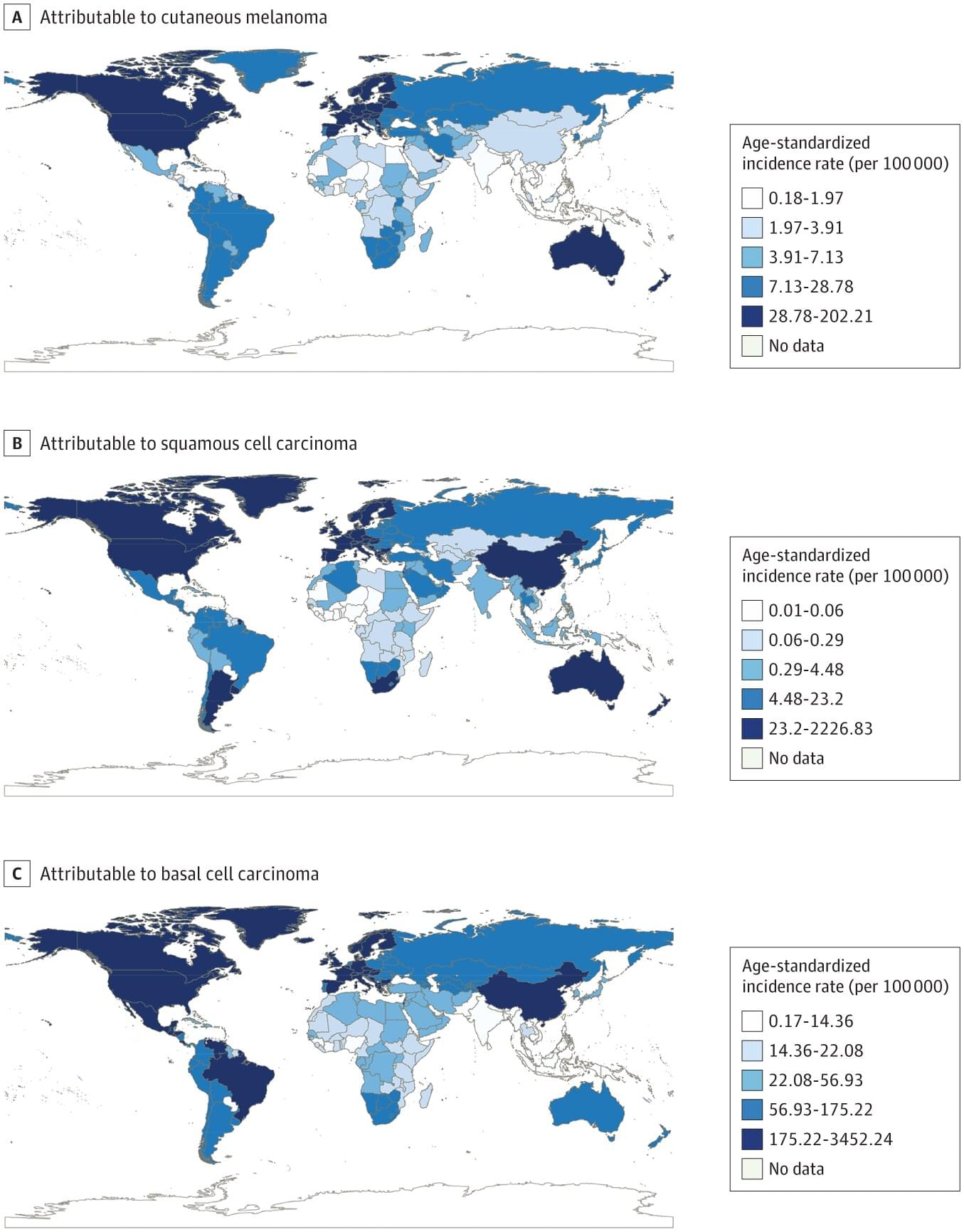
In the study, “Burden of Skin Cancer in Older Adults From 1990 to 2021 and Modelled Projection to 2050,” published in JAMA Dermatology, researchers mined the Global Burden of Diseases 2021 registry to quantify how melanoma, squamous cell carcinoma, and basal cell carcinoma affect adults aged 65 and older worldwide.