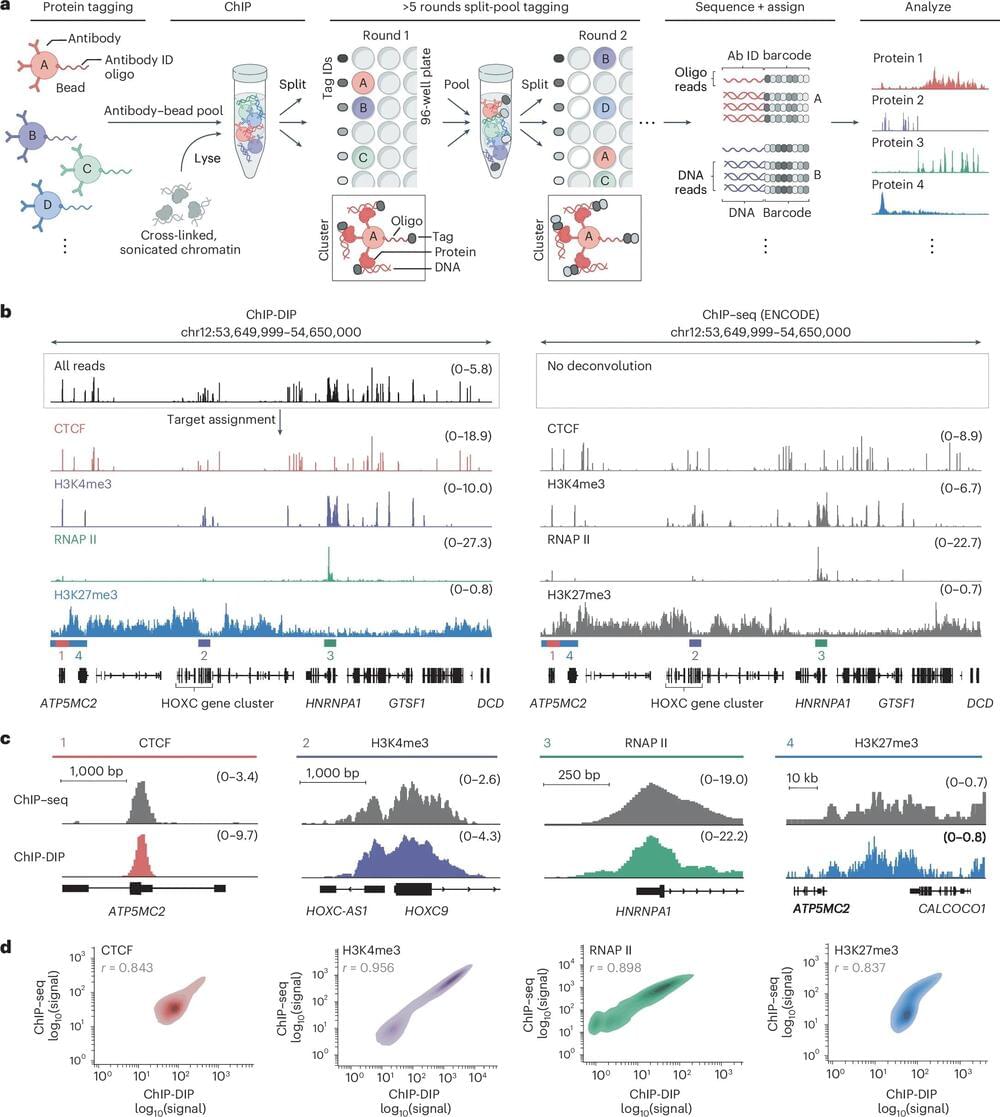
Caltech researchers have developed a new method to map the positions of hundreds of DNA-associated proteins within cell nuclei all at the same time. The method, called ChIP–DIP (Chromatin ImmunoPrecipitation Done In Parallel), is a versatile tool for understanding the inner workings of the nucleus during different contexts, such as disease or development.
The research was conducted in the laboratory of Mitchell Guttman, professor of biology, and is described in a paper that appears in the journal Nature Genetics.
Nearly all cells in the human body contain the same DNA, which encodes the blueprint for creating every cell type in the body and directing their activities. Despite having the same genetic material, different cell types express unique sets of proteins, allowing for the various cells to perform their specialized functions and to adapt to conditions within their environments. This is possible because of careful regulation within the nucleus of each cell and involves thousands of regulatory proteins that localize to precise places in the nucleus.