Experiments have shown how the world’s hardiest microbe could endure freezing, dry and irradiated conditions on Mars.



Thanks to NASA, the world may soon have access to chargers that can top off an EV in as little as five minutes. One of the biggest obstacles to fast charging is dealing with temperature. According to NASA, for an EV to be charged in five minutes, the charger must deliver an electric current of 1,400 amperes. For reference, the fastest chargers currently available max out at around 520 amperes. More amperes equals more heat. A lot more heat. Companies and research organizations are pursuing solutions to the problem; Ford and Purdue University, for example, are exploring liquid-cooled charging cables.
A team sponsored by NASA’s Biological and Physical Sciences Division is working on technology that could provide another solution needed for ultra fast EV charging. The technology has been developed for use in space, in which massive temperature differentials require massive heat transfer capabilities. An experiment to prove the new tech, the Flow Boiling and Condensation Experiment (FBCE), was installed on the International Space Station and is providing data that NASA will use to determine if the system will provide the claimed orders-of-magnitude benefits in heat transfer efficiency.
We’re definitely not NASA-level engineers but we will try to explain the FBCE the best we can. The FBCE is made up of several modules; one of which is called a “Flow Boiling Module” (FBM). When cooling liquid inside the FBM begins to boil, the bubbles formed draw liquid from the inner part of the flow channel to its walls. The process “efficiently transfers heat by taking advantage of both the liquid’s lower temperature and the ensuing change of phase from liquid to vapor.” The technique has been dubbed “subcooled flow boiling.”

An international research team led by the Department of Microstructured Quantum Matter at the MPSD reports the first observation of switchable chiral transport in a structurally achiral crystal, the Kagome superconductor CsV3Sb5. Their work has been published in Nature.
Whether or not an object is indistinguishable from its mirror image has important consequences for its physical behavior. Say you watch a basketball player in a mirror. The ball, the player and their surroundings are, at first glance, just the same in the mirror as in real life. But if observed closely, some details are different. The ball in the player’s right hand now appears in their left hand in the mirror. While the mirror image still shows the same hand, it has clearly changed from a left to a right hand or vice versa. Many other physical objects also have mirror images that differ in a key aspect, just like hands, which is why scientists call them handed or chiral (from Greek χϵρι = hand). Others, like the ball, cannot be distinguished from their mirror image, which makes them achiral.
Chirality is one of the most fundamental geometric properties and plays a special role in biology, chemistry and physics. It can cause surprising effects: One version of the carvone molecule, for example, produces a spearmint smell but its chiral—mirrored—equivalent smells of caraway.

Neuroscience is contributing to an understanding of the biological bases of human intelligence differences. This work is principally being conducted along two empirical fronts: genetics—quantitative and molecular—and brain imaging. Quantitative genetic studies have established that there are additive genetic contributions to different aspects of cognitive ability—especially general intelligence—and how they change through the lifespan. Molecular genetic studies have yet to identify reliably reproducible contributions from individual genes. Structural and functional brain-imaging studies have identified differences in brain pathways, especially parieto-frontal pathways, that contribute to intelligence differences. There is also evidence that brain efficiency correlates positively with intelligence.
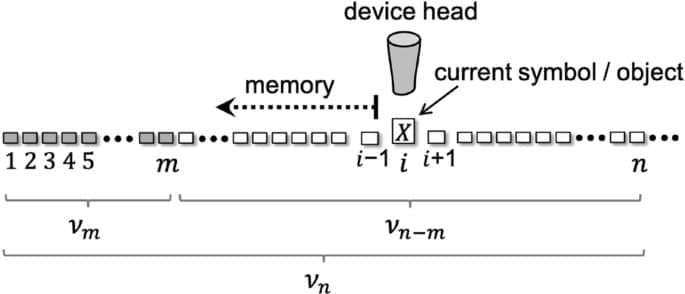
Information variations in a chain-like system are associated to energy transactions with the environment, which can take place reversibly or irreversibly, with a lower theoretical energy limit22,23. Fluctuations as a consequence of pure computations are on the order of the thermal level (i.e., similar to kT, being k the Boltzmann constant and T the absolute temperature), according to Landauer’s principle. Such energies are negligible at routine human scales but become significant when the size of the system is nanoscopic or smaller, because the work and heat it generates also compare with the thermal level. Small systems are based on nanostructures, including individual molecules and arrangements of atoms, such as biological and quantum systems.
Fluctuation theorems have appeared in recent years explaining quantitatively energy imbalances between forward and reverse pathways or between equilibrium and non-equilibrium processes24,25. They have been tested experimentally26,27,28, mostly in biomolecular systems analyzed on a one-by-one basis29. Most of these theorems establish relations among thermodynamic potentials for general systems, often with no specific insight into information theory. This theory, in turn, deals with spatially-indexed, 1-dimensional arrangements of symbols, which may not be necessarily associated to a time order. Recent generalizations separate the role of information and feedback control30,31, but still the interpretation of non-Markovianity, irreversibility and reversibility in terms of purely informational operations such as reading, writing and error correction32,33 remains obscured.
Here, we analyze energy exchanges associated to the symbolic management of a sequence of characters, without reference to the physical construction of the chain. Just by considering reversibility at the single sequence level and conservation laws, we next present two pairs of fluctuations equalities in the creation of information sequences, which use depends on energy exchange constraints. Our analysis integrates key information concepts, namely, reading, writing, proof reading and editing in the thermodynamic description of a string of symbols with information.
MR1805/iStock.
The study was published in Nature Astronomy, and it details how simple microbes that fed on hydrogen and excreted methane were likely abundant on Mars roughly 3.7 billion years ago. This was at approximately the same time that the earliest life was forming in Earth’s oceans.

Michael Levin is a developmental and synthetic biologist at Tufts University, where he is the Vannevar Bush Distinguished Professor of biology. He is a director of the Allen Discovery Center, director at the Tufts Center for Regenerative and Developmental Biology, and principal investigator at the Levin Lab.
0:00 intro.
1:38 bioelectricity and developmental biology.
7:56 memory and conditioning in GRNs.
11:50 is there a privileged cognitive substrate?
13:55 Godel type limits.
15:45 multi-scale competency architecture.
25:12 intelligence.
27:00 conceptual framework for cognition.
29:45 does cognition bottom out somewhere?
36:47 synthetic cognition.
39:23 sci-fi that captures this well.
45:16 consciousness, hard problem, and the consciousness of development.
51:09 where does the self come from?
54:06 how do different emergent levels interact.
56:50 top-down causality.
1:02:28 where do goals come from?
1:07:06 balancing conceptual and empirical work.
Michael Levin’s Website:
https://ase.tufts.edu/biology/labs/levin/
Podcast.
Spotify: https://open.spotify.com/show/0dUBLTl6qzOfA0xMndLFzq.
Google: https://www.google.com/podcasts?feed=aHR0cHM6Ly9mZWVkcy5yZWR…FhMw%3D%3D
Apple: https://podcasts.apple.com/us/podcast/thing-in-it-self/id1616881426
Amazon: https://music.amazon.ca/podcasts/9c6c08b2-e975-47d6-a897…in-it-self.
Social.
Twitter: https://twitter.com/thinginitself__
Instagram: https://www.instagram.com/thinginitself.pod/
A paradigm shift in how we think about the functions of the human brain. A long-awaited genetic sequence of Rafflesia arnoldii, the strangest flower in the world. A revelation in sleep science. These are some of the year’s biggest discoveries in neuroscience and other areas of biology. Read the articles in full at Quanta: https://www.quantamagazine.org/the-year-in-biology-20211221/
Quanta Magazine is an editorially independent publication supported by the Simons Foundation.