:oooooo.
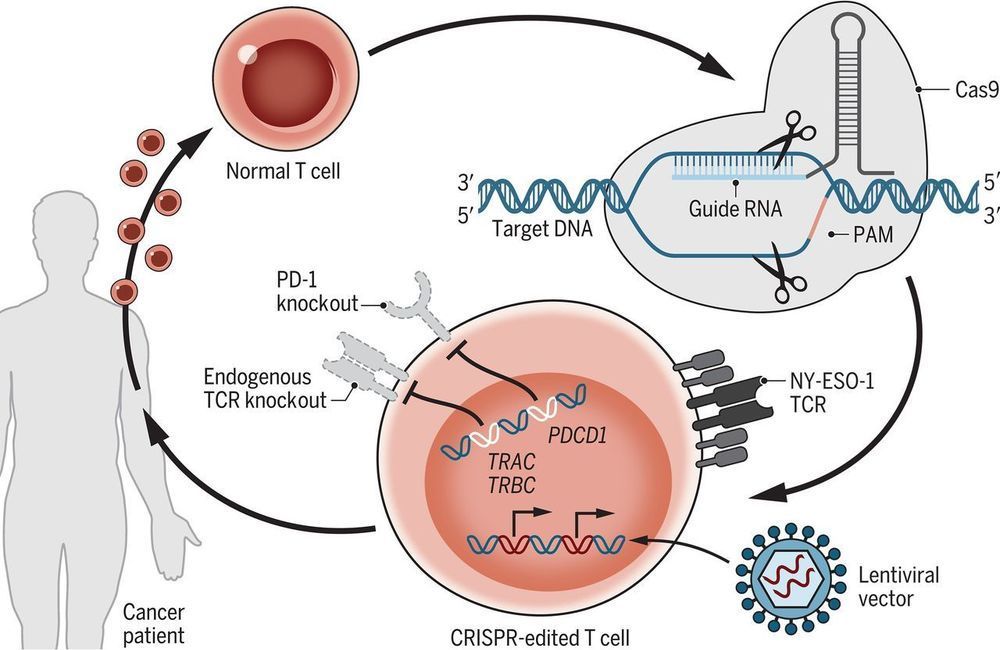
CRISPR-Cas9 is a revolutionary gene-editing technology that offers the potential to treat diseases such as cancer, but the effects of CRISPR in patients are currently unknown. Stadtmauer et al. report a phase 1 clinical trial to assess the safety and feasibility of CRISPR-Cas9 gene editing in three patients with advanced cancer (see the Perspective by Hamilton and Doudna). They removed immune cells called T lymphocytes from patients and used CRISPR-Cas9 to disrupt three genes (TRAC, TRBC, and PDCD1) with the goal of improving antitumor immunity. A cancer-targeting transgene, NY-ESO-1, was also introduced to recognize tumors. The engineered cells were administered to patients and were well tolerated, with durable engraftment observed for the study duration. These encouraging observations pave the way for future trials to study CRISPR-engineered cancer immunotherapies.