A new software tool, ovrlpy, improves quality control in spatial transcriptomics, a key technology in biomedical research. Developed by the Berlin Institute of Health at Charité (BIH) in international collaboration, ovrlpy is the first tool to identify cell overlaps and folds in tissue sections, thereby reducing previously unrecognized sources of misinterpretations. The researchers have published their results in the journal Nature Biotechnology.
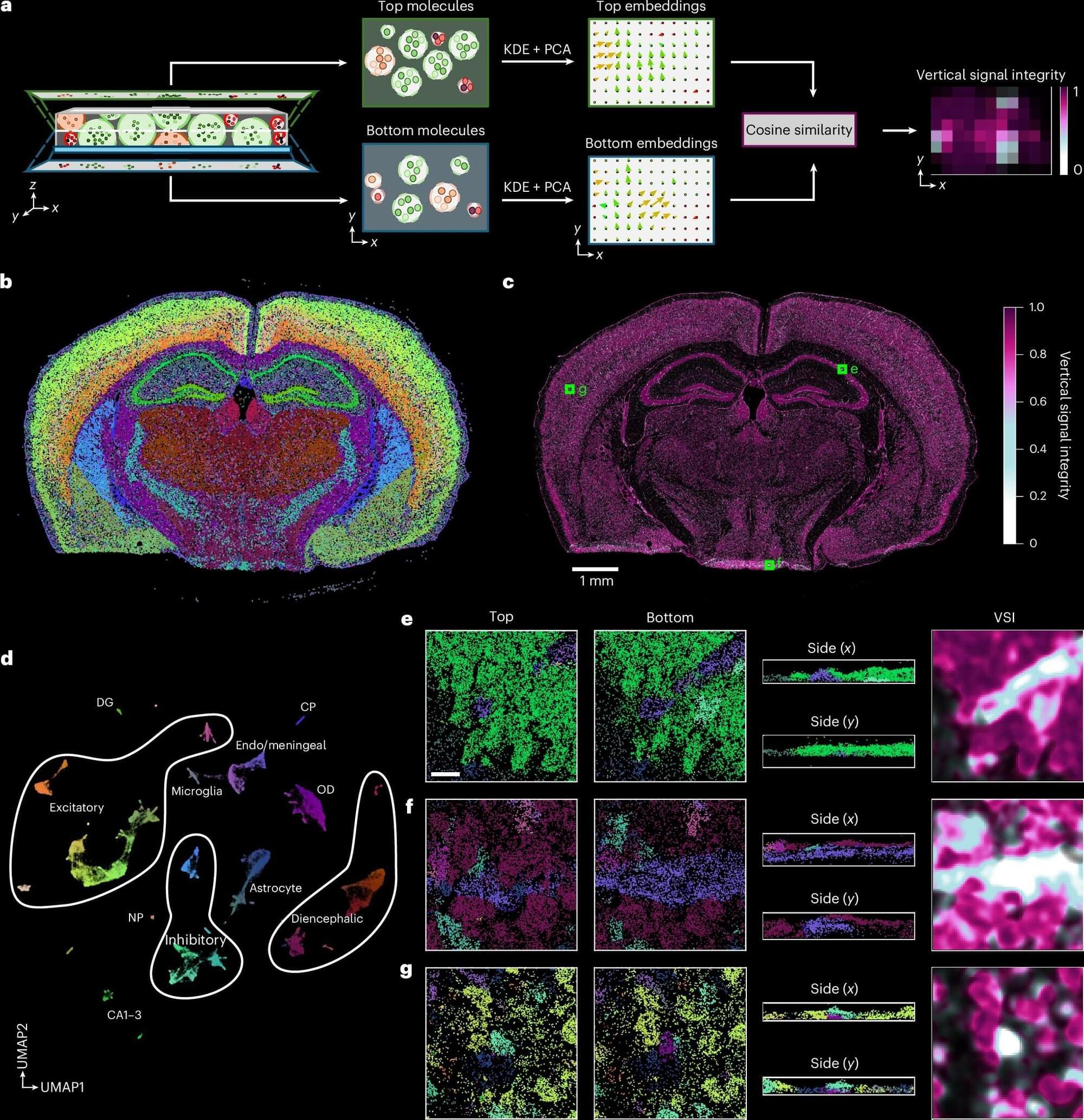
Spatial transcriptomics is a pioneering field of research in biomedicine that visualizes cellular activity within a tissue by mapping RNA transcripts and assigning this molecular activity to individual cells. So far, such analyses of tissue samples have mostly been interpreted in two dimensions. However, even very thin tissue sections of 5 to 10 micrometers thick, about one-tenth the width of a human hair, have a complex three-dimensional structure.
If this 3D arrangement is interpreted only as a flat surface, analytical errors can occur, for example, due to cell overlaps or tissue folds. This impedes the precise assignment of transcripts to individual cells and can distort downstream analysis and interpretation.