Newlypublished by gennady verkhivker, et al.
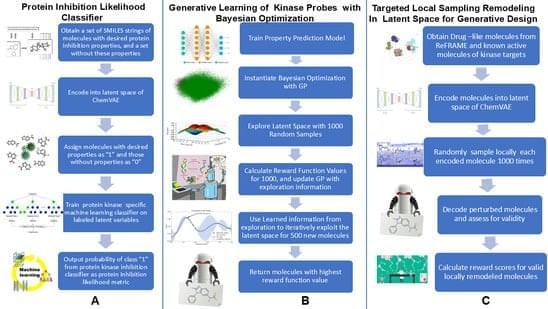
🔍 Key findings: Novel generative framework integrates ChemVAE-based latent space modeling with chemically interpretable structural similarity metric (Kinase Likelihood Score) and Bayesian optimization for SRC kinase ligand design, demonstrating kinase scaffolds spanning 37 protein kinase families spontaneously organize into low-dimensional manifold with chemically distinct carboxyl groups revealing degeneracy in scaffold encoding — local sampling successfully converts scaffolds from other kinase families into novel SRC-like chemotypes accounting for ~40% of high-similarity cutoffs.
Read now ➡️
Scaffold-aware artificial intelligence (AI) models enable systematic exploration of chemical space conditioned on protein-interacting ligands, yet the representational principles governing their behavior remain poorly understood. The computational representation of structurally complex kinase small molecules remains a formidable challenge due to the high conservation of ATP active site architecture across the kinome and the topological complexity of structural scaffolds in current generative AI frameworks. In this study, we present a diagnostic, modular and chemistry-first generative framework for design of targeted SRC kinase ligands by integrating ChemVAE-based latent space modeling, a chemically interpretable structural similarity metric (Kinase Likelihood Score), Bayesian optimization, and cluster-guided local neighborhood sampling.