A team of engineers at the University of California San Diego is making it easier for researchers from a broad range of backgrounds to understand how different species are evolutionarily related, and support the transformative biological and medical applications that rely on these species trees. The researchers developed a scalable, automated and user-friendly tool called ROADIES that allows scientists to infer species trees directly from raw genome data, with less reliance on the domain expertise and computational resources currently required.
Species trees are critical to solidifying our understanding of how species evolved on a broad scale, but can also help find functional regions of the genome that could serve as drug targets; link physical traits to genomic changes; predict and respond to zoonotic outbreaks; and even guide conservation efforts.
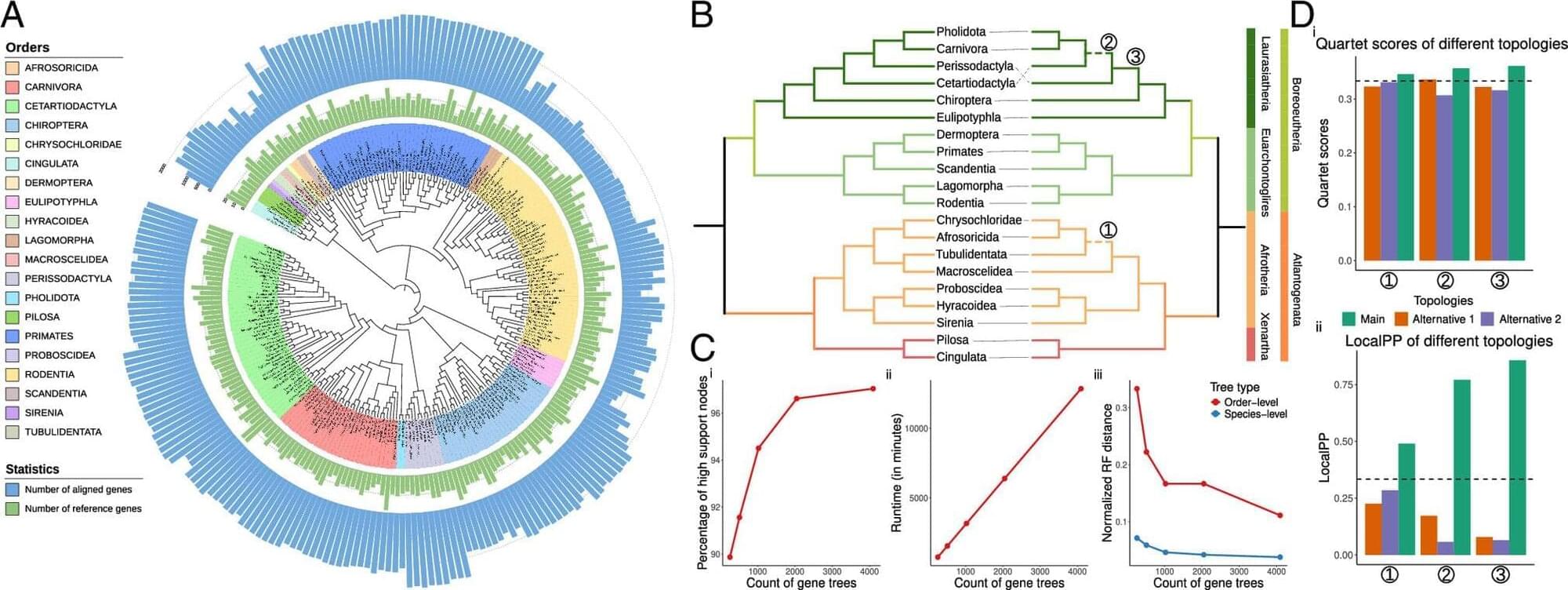
In a new paper published in the journal Proceedings of the National Academy of Sciences on May 2, the researchers, led by UC San Diego electrical and computer engineering professor Yatish Turakhia, showed that ROADIES infers species trees that are comparable in quality with the state-of-the-art studies, but in a fraction of the time and effort. This paper focused on four diverse life forms— placental mammals, pomace flies, birds and budding yeasts—though ROADIES can be used for any species.