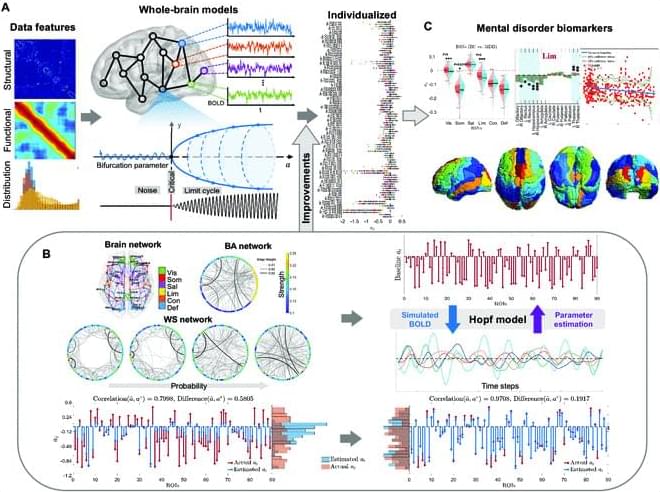
The Hopf whole-brain model, based on structural connectivity, overcomes limitations of traditional structural or functional connectivity-focused methods by incorporating heterogeneity parameters, quantifying dynamic brain characteristics in healthy and diseased states. Traditional parameter fitting techniques lack precision, restricting broader use. To address this, we validated parameter fitting methods using simulated networks and synthetic models, introducing improvements such as individual-specific initialization and optimized gradient descent, which reduced individual data loss. We also developed an approximate loss function and gradient adjustment mechanism, enhancing parameter fitting accuracy and stability.