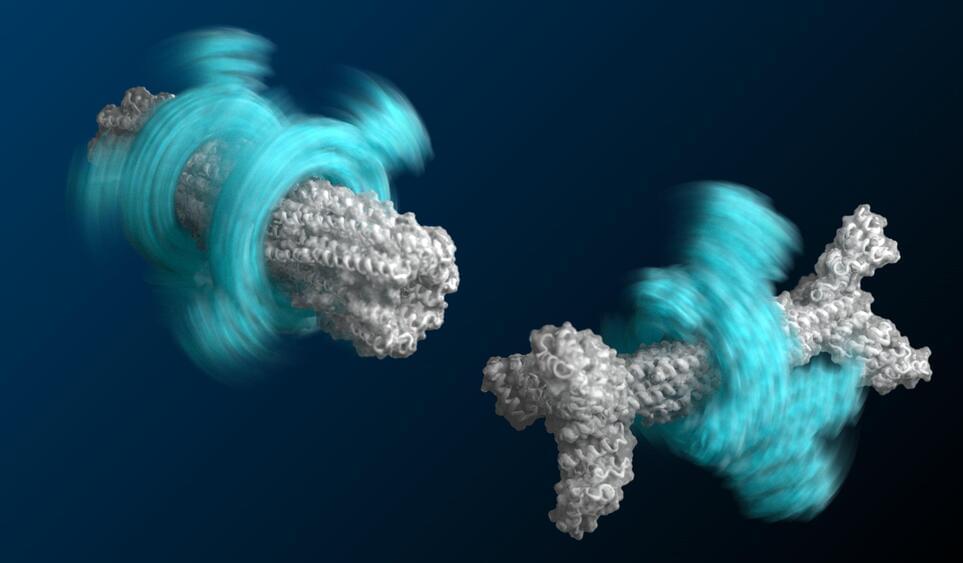
A large team of researchers at the University of Washington, working with colleagues from Université Montpellier and the Fred Hutchinson Cancer Research Center, has taken a major step toward the creation of an axle-rotor nanomachine. In their paper published in the journal Science, the group describes how they used DNA coding to customize E. coli to push them into creating proteins that assembled into rotors and axles.
As the researchers note, molecular engines are abundant in nature, from the tails of flagellum on some bacteria to the F1 motor of ATPase. And while such examples have served as good models, attempts to harness them in nature or to create new ones in the lab have been mostly unsuccessful. This is due to the single purpose features of natural engines and the unpredictability of protein folding in synthetic attempts. In this new effort, the researchers have overcome some of the hurdles that others have faced and have taken a major step toward the creation of a molecular engine by creating two of the main parts necessary for such a device—an axle and a rotor—and even managed to connect them to each other.
To create their engine parts, the researchers first used a software program called Rosetta that allowed them to design ring-like proteins with specified diameters. They then used the data from the program to add DNA coding to amino acids in E. coli bacteria that make up proteins. Such proteins are made of chains of the amino acids—it is the sequence of them that defines the shape they will take when they spontaneously fold. The team was able to coax some of the proteins into folding into rotor shapes and others into axle shapes. They then went further by coaxing multiple proteins to fold together into rotor-axle combinations—the rudimentary parts necessary for a molecular engine.