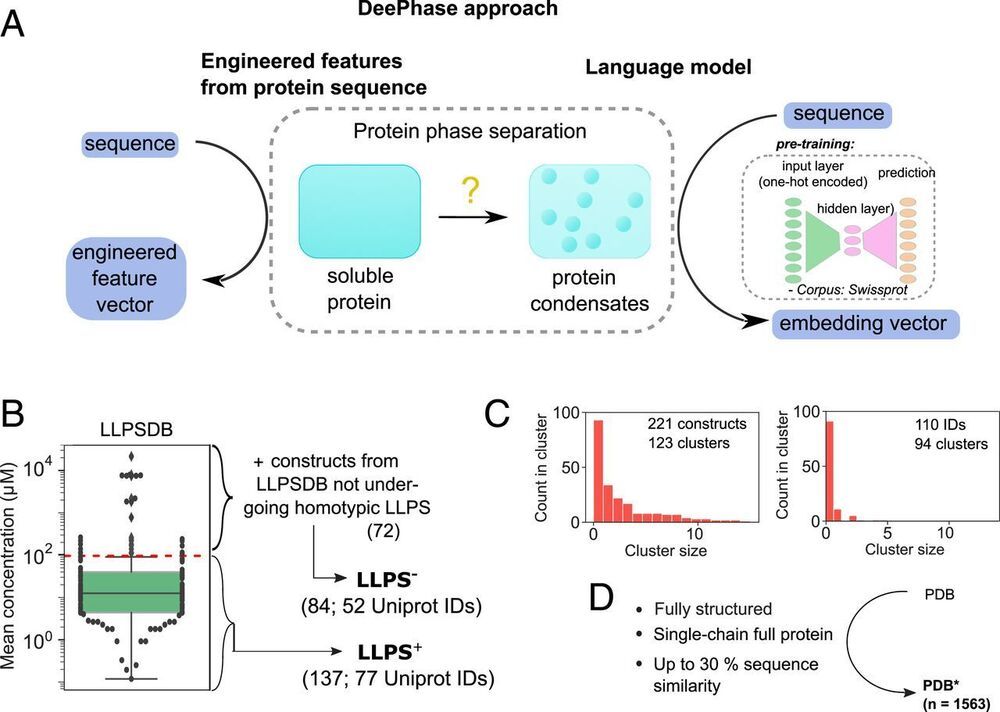
The tendency of many cellular proteins to form protein-rich biomolecular condensates underlies the formation of subcellular compartments and has been linked to various physiological functions. Understanding the molecular basis of this fundamental process and predicting protein phase behavior have therefore become important objectives. To develop a global understanding of how protein sequence determines its phase behavior, we constructed bespoke datasets of proteins of varying phase separation propensity and identified explicit biophysical and sequence-specific features common to phase-separating proteins. Moreover, by combining this insight with neural network-based sequence embeddings, we trained machine-learning classifiers that identified phase-separating sequences with high accuracy, including from independent external test data.
Intracellular phase separation of proteins into biomolecular condensates is increasingly recognized as a process with a key role in cellular compartmentalization and regulation. Different hypotheses about the parameters that determine the tendency of proteins to form condensates have been proposed, with some of them probed experimentally through the use of constructs generated by sequence alterations. To broaden the scope of these observations, we established an in silico strategy for understanding on a global level the associations between protein sequence and phase behavior and further constructed machine-learning models for predicting protein liquid–liquid phase separation (LLPS). Our analysis highlighted that LLPS-prone proteins are more disordered, less hydrophobic, and of lower Shannon entropy than sequences in the Protein Data Bank or the Swiss-Prot database and that they show a fine balance in their relative content of polar and hydrophobic residues.