Amid mounting concern about a novel coronavirus spreading from China, Lawrence Livermore National Laboratory (LLNL) researchers have developed a preliminary set of predictive 3D protein structures of the virus to aid research efforts to combat the disease.
The models are based on the genomic sequence of the novel coronavirus and a protein found in the virus that causes Severe Acute Respiratory Syndrome (SARS), which closely resembles the new virus.
The researchers plan to use the models to accelerate countermeasure design, using a combination of machine learning, biological experiments and simulation on supercomputers.
As global concern continues to rise about a novel coronavirus spreading from China, a team of Lawrence Livermore National Laboratory (LLNL) researchers has developed a preliminary set of predictive 3D protein structures of the virus to aid research efforts to combat the disease.
The team’s predicted 3D models, developed over the past week using a previously peer-reviewed modeling process, are based on the genomic sequence of the novel coronavirus and the known structure of a protein found in the virus that causes Severe Acute Respiratory Syndrome (SARS), also a coronavirus that closely resembles the new virus.
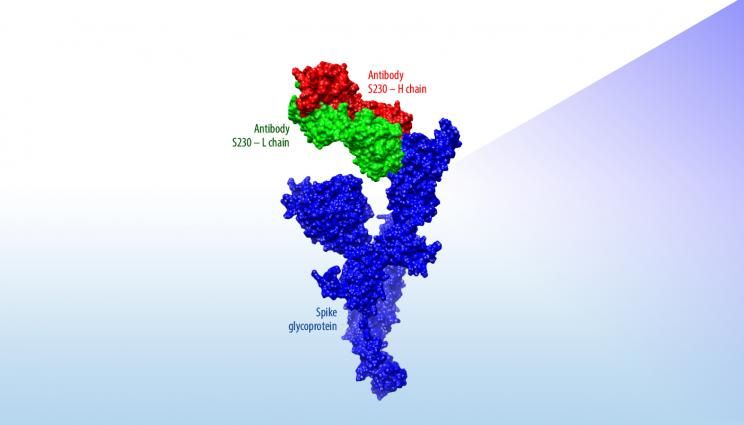
“A major part of the value of these new structural models is that they present the predicted protein in complex with SARS-neutralizing antibodies,” said Adam Zemla, an LLNL structural biologist and mathematician. “This can be thought of as the first step for the global research community to identify and model how therapeutic antibodies can be designed to fight the novel coronavirus.”