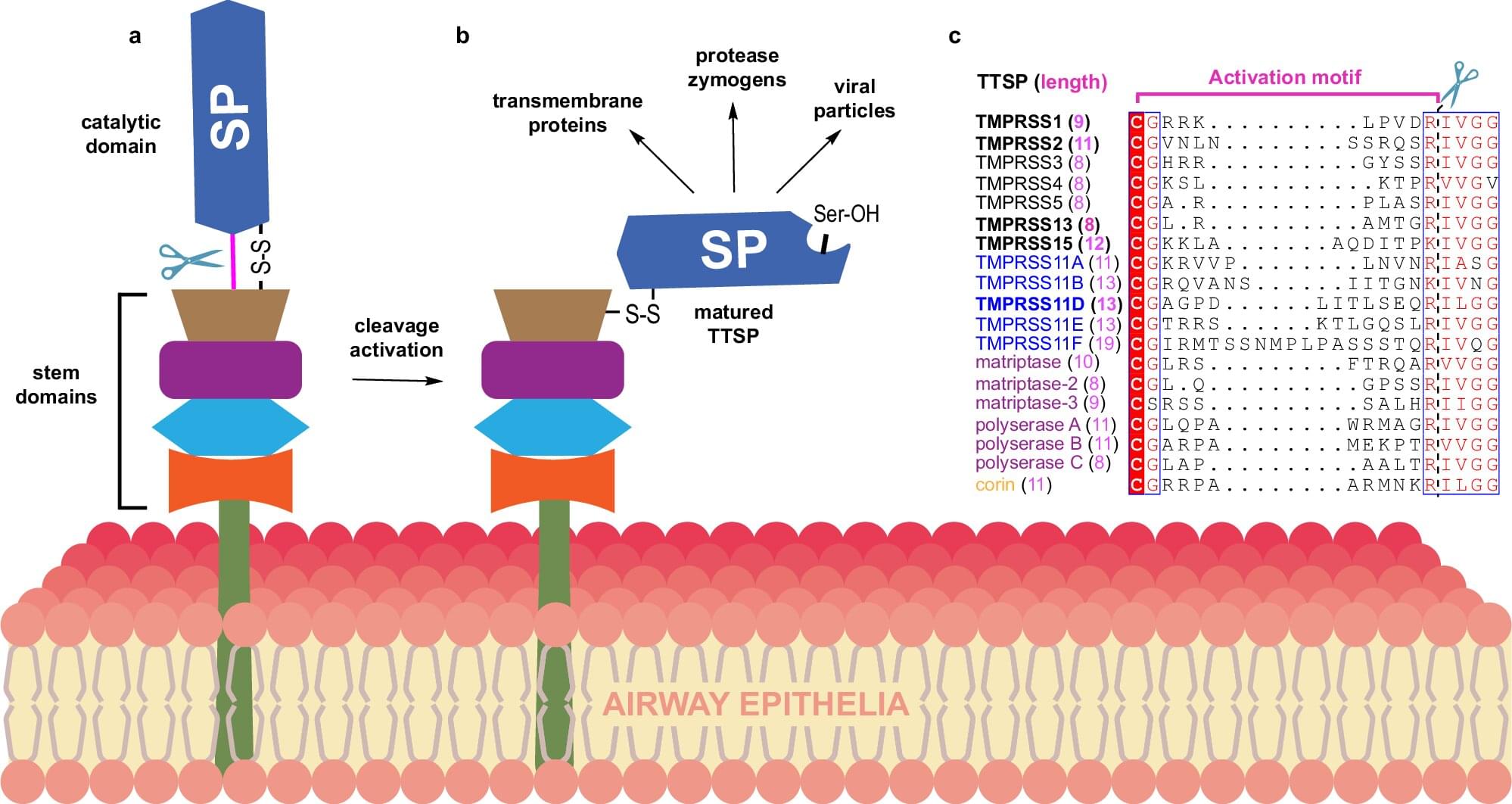
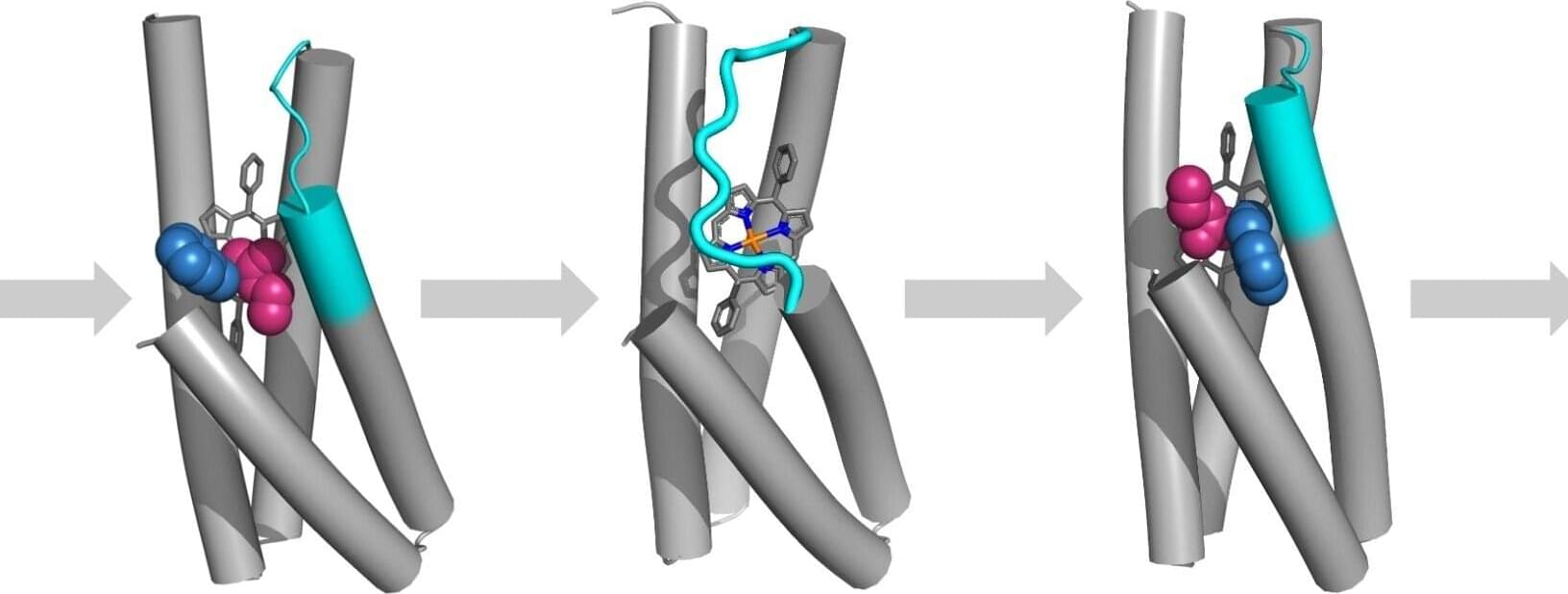
New research by scientists at the University of Toronto and the Structural Genomics Consortium has deepened our understanding of how viruses like the flu, common cold, and COVID-19 get into cells in human airways.
Using the Canadian Light Source at the University of Saskatchewan, the researchers identified for the first time the crystal structures of a human protein (TMPRSS11D) that viruses use as a doorway into our body. The study is published in the journal Nature Communications.
Understanding how viruses use our proteins to gain entry into our cells will help researchers develop better ways to stop infections in their tracks.