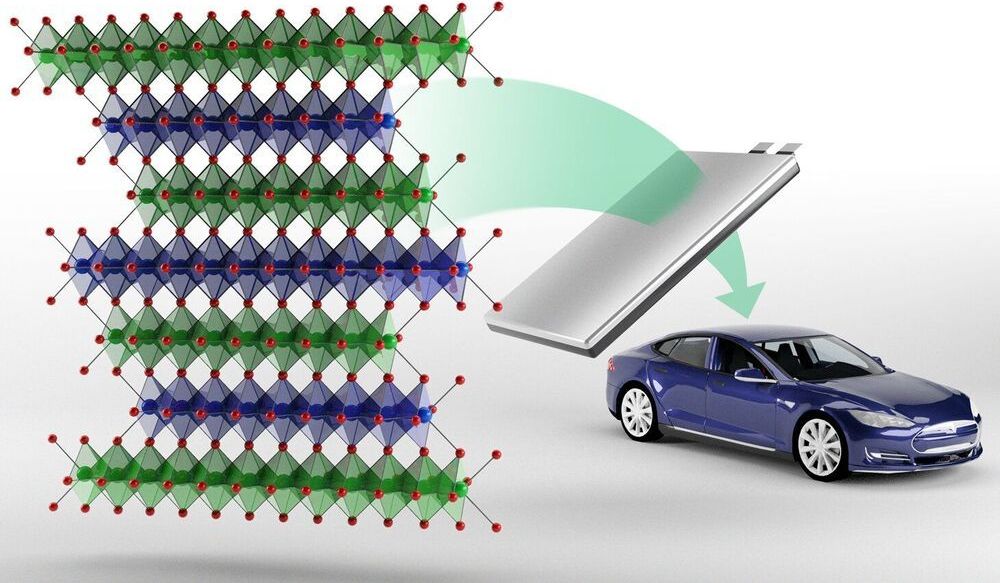
Oak Ridge National Laboratory researchers have developed a new family of cathodes with the potential to replace the costly cobalt-based cathodes typically found in today’s lithium-ion batteries that power electric vehicles and consumer electronics.
The new class called NFA, which stands for nickel-, iron-and aluminum-based cathode, is a derivative of lithium nickelate and can be used to make the positive electrode of a lithium-ion battery. These novel cathodes are designed to be fast charging, energy dense, cost effective, and longer lasting.
With the rise in the production of portable electronics and electric vehicles throughout the world, lithium-ion batteries are in high demand. According to Ilias Belharouak, ORNL’s scientist leading the NFA research and development, more than 100 million electric vehicles are anticipated to be on the road by 2030. Cobalt is a metal currently needed for the cathode which makes up the significant portion of a lithium-ion battery’s cost.