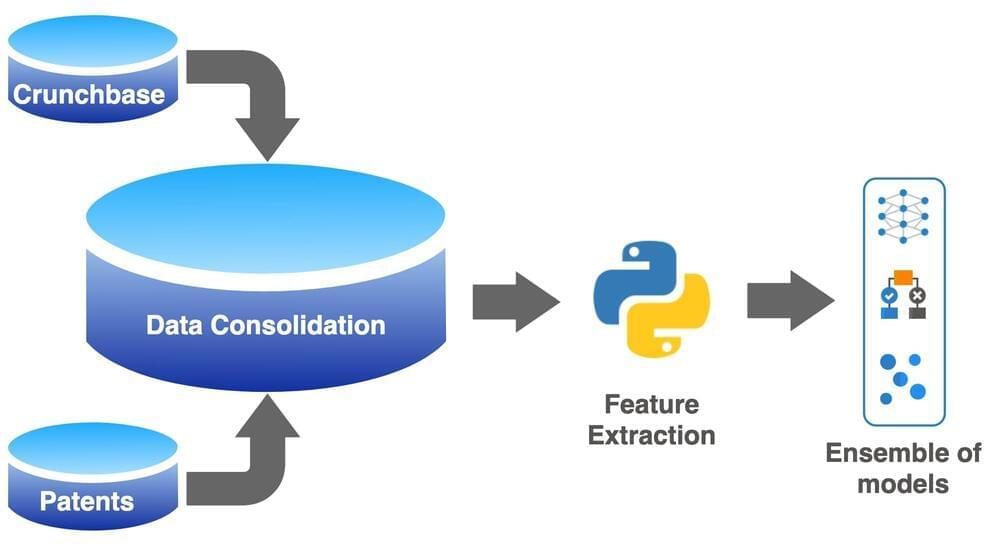
A study in which machine-learning models were trained to assess over 1 million companies has shown that artificial intelligence (AI) can accurately determine whether a startup firm will fail or become successful. The outcome is a tool, Venhound, that has the potential to help investors identify the next unicorn.
It is well known that around 90% of startups are unsuccessful: Between 10% and 22% fail within their first year, and this presents a significant risk to venture capitalists and other investors in early-stage companies. In a bid to identify which companies are more likely to succeed, researchers have developed machine-learning models trained on the historical performance of over 1 million companies. Their results, published in KeAi’s The Journal of Finance and Data Science, show that these models can predict the outcome of a company with up to 90% accuracy. This means that potentially 9 out of 10 companies are correctly assessed.
“This research shows how ensembles of non-linear machine-learning models applied to big data have huge potential to map large feature sets to business outcomes, something that is unachievable with traditional linear regression models,” explains co-author Sanjiv Das, Professor of Finance and Data Science at Santa Clara University’s Leavey School of Business in the US.