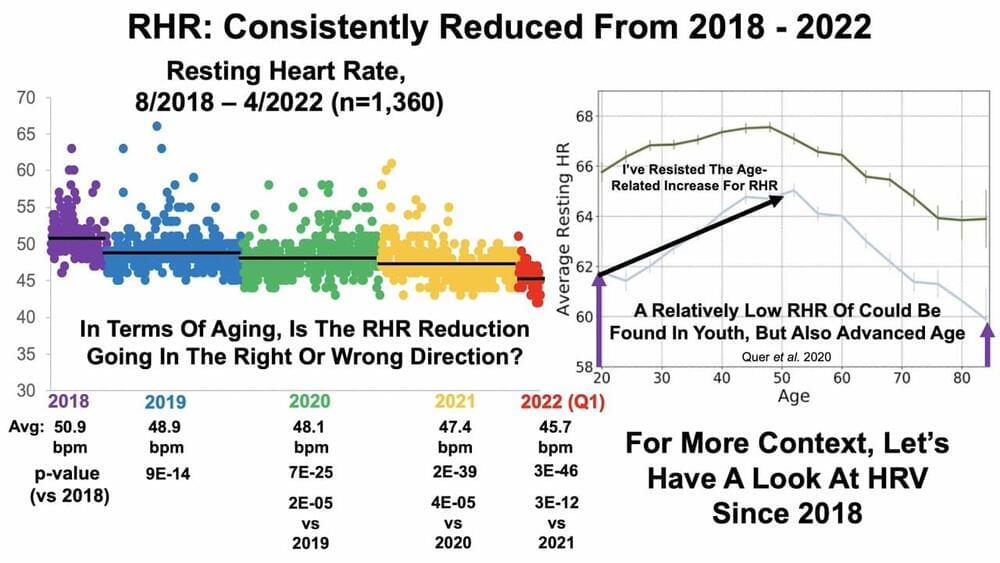
This just came out, a day or so ago.
Can the aging process be reversed — or even halted, altogether? If we manage to decode this final mystery of our human biology, we might soon be able to eradicate age-related illnesses like cancer, dementia and heart problems.
The race to invent the miracle pill is well underway. Today, international researchers are getting astonishingly close to realizing humanity’s dream of immortality.
The hunt for immortality gained traction with the discovery of Costa Rica’s so-called “Blue Zone, by Luis Rosero-Bixby. In the “Blue Zone, on the Nicoya Peninsular, he found a remarkable number of centenarians. Here, male life expectancy is the highest in the world. Their healthy lifestyle is one factor, but the promise of longevity is probably also because their telomeres — sections of DNA found at the end of chromosomes — are longer than those of the average person.
It’s a field of research currently being explored by Maria Blasco in Madrid. But this is just one of many possible factors influencing the process of aging. Senescent cells may also play a key role. Also known as “zombie cells, these attack our body in old age and flood it with alarm signals until, at some point, we collapse under their weight. That’s a theory proposed by another researcher in Spain, Manuel Serrano.