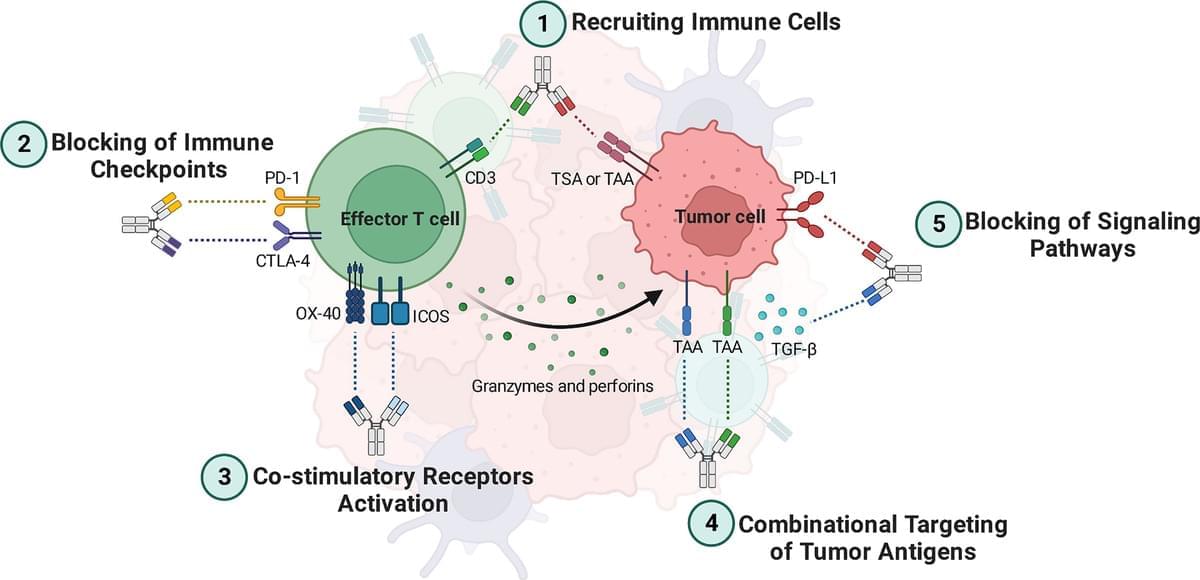
Antibody-based cancer immunotherapy has become a powerful asset in the arsenal against malignancies. In this regard, bispecific antibodies (BsAbs) are a grou…

My new AI-assisted short film is here. Kira explores human cloning and the search for identity in today’s world.
It took nearly 600 prompts, 12 days (during my free time), and a $500 budget to bring this project to life. The entire film was created by one person using a range of AI tools, all listed at the end.
Enjoy.
~ Hashem.
Instagram: / hashem.alghaili.
Facebook: / sciencenaturepage.
Other channels: https://muse.io/hashemalghaili
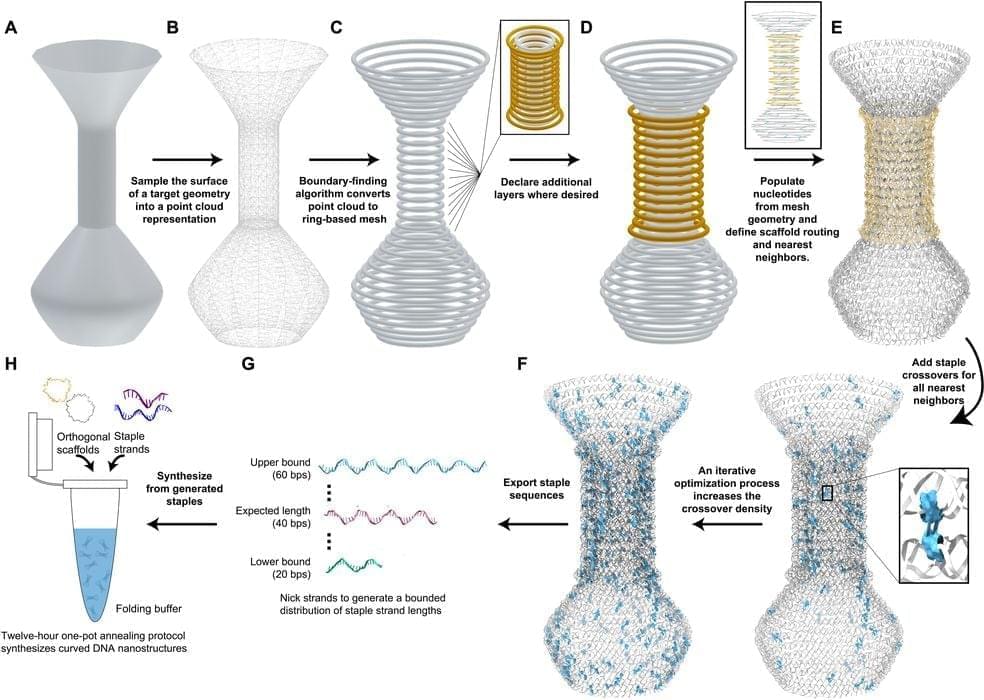
A research team is working on ways to produce 3D structures using DNA.
DNA is the foundation of life, as DNA molecules carry digital information that is used by cells to reproduce and develop all the functional biology required for life.
DNA can do this because it contains four molecules that can be easily bonded into complex structures. The molecules, adenine (A), cytosine ©, guanine (G), and thymine (T), can be arranged in “ladder” forms that are self-assembling.


Wimbledon’s new automated line-calling system glitched during a tennis match Sunday, just days after it replaced the tournament’s human line judges for the first time.
The system, called Hawk-Eye, uses a network of cameras equipped with computer vision to track tennis balls in real-time. If the ball lands out, a pre-recorded voice loudly says, “Out.” If the ball is in, there’s no call and play continues.
However, the software temporarily went dark during a women’s singles match between Brit Sonay Kartal and Russian Anastasia Pavlyuchenkova on Centre Court.

These black holes were whirling at speeds nearly brushing the limits of Einstein’s theory of general relativity, forcing researchers to stretch existing models to interpret the signal.
Breathtaking images of gamma-ray flare from supermassive black hole M87
“Black holes this massive are forbidden by standard stellar evolution models,” says Professor Mark Hannam from Cardiff University. “One explanation is that they were born from past black hole mergers, a cosmic case of recursion.”



Why is Mars barren and uninhabitable, while life has always thrived here on our relatively similar planet Earth?
A discovery made by a NASA rover has offered a clue for this mystery, new research said Wednesday, suggesting that while rivers once sporadically flowed on Mars, it was doomed to mostly be a desert planet.
Mars is thought to currently have all the necessary ingredients for life except for perhaps the most important one: liquid water.