Senior corporate officers across the nation are wringing their hands. The 2023 economic climate is uncertain, but one thing is for sure—more layoffs are coming. In November 2022 alone, more than 80,000 layoffs were announced from tech giants like Meta, Amazon, and Twitter, as well as conventional companies like PepsiCo, Goldman Sachs, and Ford.
Downsizing is one of the most difficult things that leaders ever have to accomplish. How many should you let go? When should you do it? Who stays and who goes? What severance do you offer? How do you protect your diversity targets? How to maintain trust and productivity in those who get to stay?
Let too many go, too fast, and you could damage service and execution. Let too few go, too late, and you might lose money. Let the wrong people go and you may just create internal chaos. Getting this wrong can have enormous consequences on profitability, productivity, brand reputation, and stock price.
A company has donated 350 of Smit’s, the biomechanical engineer behind the design, 3D-printed prosthetic hands to war victims in Ukraine.
Delft University of Technology (TU Delft) researchers designed laser-cutting 3D-printed prosthetic hands for Ukranian war victims. Thanks to laser-cutting technology, war victims get their prosthetic limbs more easily. These prosthetic hands are in use in India, and Indian company Vispala donated 350 of Smit’s 3D-printed prosthetic hands to war victims in Ukraine, according to the TU Delft.
Designed by biomedical engineer Gerwin Smit, the so-called “Hundred Dollar Hand” is very cheap to produce. Smit’s artificial hand offers a sturdy and trustworthy option because 80 percent of persons who require a prosthetic hand reside in nations with few resources. Delft-produced 350 prosthetic hands donated to Ukrainian War wictims.
Nothing personal, the Sun is just going through a phase right now. On Jan 4, 2023, our planet reached the closest point to the Sun in its orbit and is expected to be hit by the wake of a coronal mass ejection (CME) coming from the Sun, Live Science reported.
IStock/cokada.
Solar activity has been picking up pace in the past few months as the Sun approaches the peak of its solar cycle. Every 11 years or so, the poles on the Sun switch their positions, sending our star into a tizzy of activity, marked by the appearance of sunspots and darker areas on the solar surface.
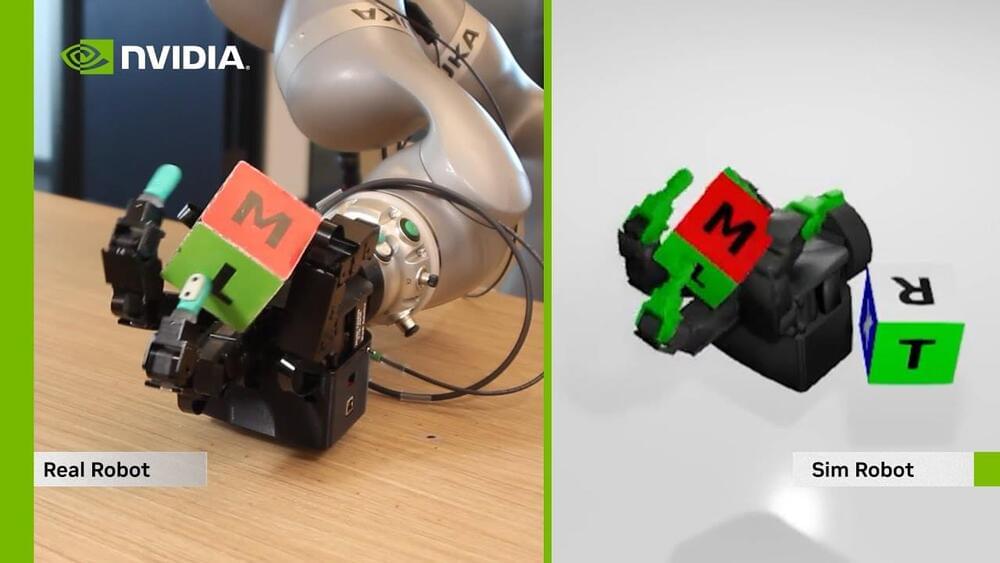
Breakthroughs in robotic manipulation will enable a new wave of applications for robots in commercial, industrial and home applications.
NVIDIA’s Isaac Sim and Gym tools allow robots to be trained entirely in simulation before their AI brains are transferred into real robots. Our video shows the results of this sim-to-real transfer.
This result has been made possible by simulating thousands of robots in parallel using NVIDIA GPUs, and by generating millions of training images using our Omniverse Replicator tools.
https://developer.nvidia.com/isaac-sim.
https://developer.nvidia.com/nvidia-omniverse-platform/replicator.
https://www.youtube.com/c/NVIDIAOmniverse
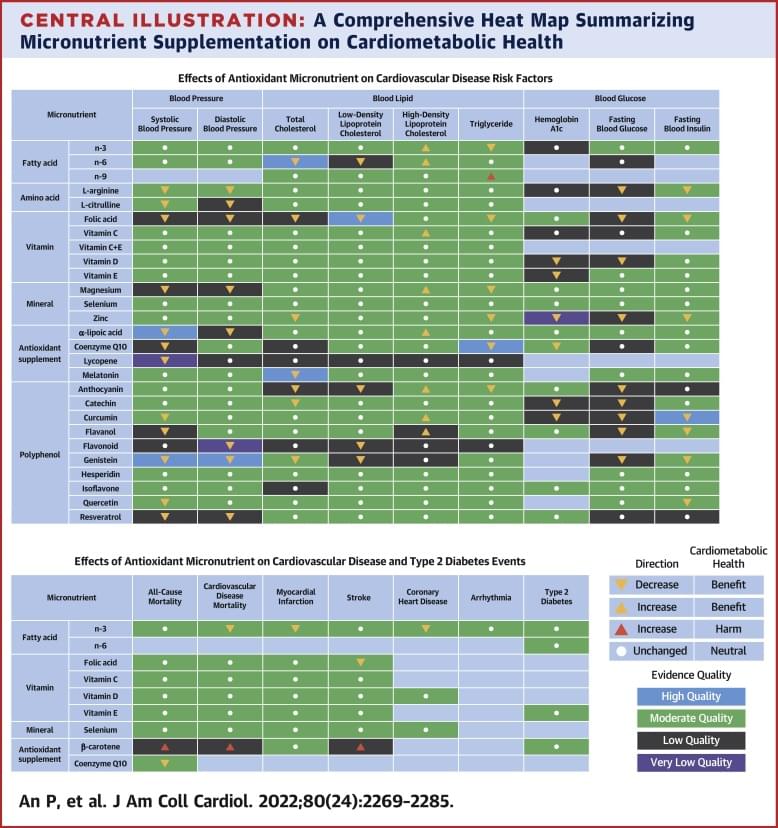
The researchers looked at randomized, controlled intervention trials evaluating 27 different types of antioxidant supplements. They found strong evidence that several offered cardiovascular benefits. These included omega-3 fatty acid, which decreased mortality from cardiovascular disease; folic acid, which lowered stroke risk; and coenzyme Q10, an antioxidant sometimes marketed as CoQ10, which decreased all-cause mortality. Omega-6 fatty acid, L-citrulline, L-arginine, quercetin, melatonin, curcumin, Vitamin D, zinc, magnesium, alpha-lipoic acid (ALA), catechin, flavanol, and genistein also showed evidence of reducing cardiovascular risk.
Not all supplements were beneficial. Vitamin C, Vitamin D, Vitamin E, and selenium showed no effect on long-term cardiovascular disease outcomes or type-2 diabetes risk. And beta carotene supplements increased all-cause mortality.
Healthy dietary patterns are rich in micronutrients, but their influence on cardiovascular disease (CVD) risks has not been systematically quantified.
The goal of this study was to provide a comprehensive and most up-to-date evidence-based map that systematically quantifies the impact of micronutrients on CVD outcomes.
This study comprised a systematic review and meta-analysis of randomized controlled intervention trials of micronutrients on CVD risk factors and clinical events.
Aubrey De Grey discusses the progress and potential of therapies related to his ideas on anti-aging medicine, including the four therapies that will be tested in a mouse rejuvenation trial. He also shares his thoughts on partnering with organizations and individuals in the field, integrating AI into his work, and the importance of structure in maximizing impact. Aubrey de Grey discusses the potential for Yamanaka factors to be used in organ rejuvenation, and the role of transcription factors in creating induced pluripotent stem cells. He also provides advice for those interested in getting involved in the field and shares his views on time management and productivity. Aubrey De Grey discusses the potential for reversing the pathology of aging to address mechanical issues and mentions promising research being conducted by MAIA Biotechnology on cancerous cells that express telomerase. He also expresses his optimism about the possibility of reaching “longevity escape velocity” within the next 15 years.
Youtube:
Aubrey De Grey links.
https://twitter.com/aubreydegrey?ref_src=twsrc%5Egoogle%7Ctw…r%5Eauthor.
https://www.levf.org/
https://www.linkedin.com/in/aubrey-de-grey-24260b/
PODCAST INFO:
The Learning With Lowell show is a series for the everyday mammal. In this show we’ll learn about leadership, science, and people building their change into the world. The goal is to dig deeply into people who most of us wouldn’t normally ever get to hear. The Host of the show – Lowell Thompson-is a lifelong autodidact, serial problem solver, and founder of startups.
LINKS
Youtube: https://www.youtube.com/channel/UCzri06unR-lMXbl6sqWP_-Q
Youtube clips: https://www.youtube.com/channel/UC-B5x371AzTGgK-_q3U_KfA
Linkedin: https://www.linkedin.com/in/lowell-thompson-2227b074
Twitter: https://twitter.com/LWThompson5
Website: https://www.learningwithlowell.com/
Podcast email: [email protected].
Timestamps.
00:00 Start.
01:00 Mark Hamaleinen write in question: Why we still don’t have any therapies based on his ideas published 20 years ago in his breakout paper.
04:30 update on escape velocity. 50% in 15 years.
08:30 Experiments on mice.
12:00 Yamanaka factor thoughts, current research on mice continued.
14:30 Lev foundation research expanded and explained.
16:30 Lev foundation thesis compared to others.
21:00 Hardships being at the tip of the innovation field of longevity.
23:45 Open source, non profits, lev foundation.
26:00 Ideas from previous organizations (ie. SENS) applied in LEV
27:15 Ichor therapeutics, and partnership process of LEV
29:30 Next generation mentorship.
30:30 Summer internship program.
32:45 Bullish on longevity.
33:30 AI role in longevity.
35:55 Anything fundamental making longevity therapies only for the rich.
38:30 Longevity surgery therapies.
39:30 Advice for people.
40:30 Bottlenecks of longevity.
42:00 Books!
43:05 Staying on cutting edge/ learning.
44:15 Current curiosity and fascination.
46:45 What happiness means to him and how he optimizes for it.
48:30 how he stays healthy over the years, longevity practices he uses.
51:45 How much money does he and the space need.
52:55 Anything stopping him from getting Jeff Bezos and other high network people to invest or donate.
51:55 Thoughts on altos labs & calico labs.
56:35 up and coming people that inspire him (crypto people, michael levin)
59:00 Finding up and coming people to work with and for (advice)
60:30 Things people get wrong and what people ask him about longevity.
1:01:35 Aubrey newsletter and updates (news suggestions)
1:03:30 Question he wonders about that doesn’t have answer to.
1:04:15 How he maximizes his day and stays productive.
1:05:15 Thoughts on MAIA Biotechnology, telomerase, short vs long.
1:09:30 Final thoughts on lev foundation.
#longevity #aubreydegrey #LEVF
The latest Omicron variant is making scientists take notice in the US — so what do you need to know?
The reason your three-pound brain doesn’t feel heavy is because it floats in a reservoir of cerebrospinal fluid (CSF), which flows in and around your brain and spinal cord. This liquid barrier between your brain and skull protects it from a hit to your head and bathes your brain in nutrients.
But the CSF has another critical, if less known, function: it also provides immune protection to the brain. Yet, this function hasn’t been well studied.
A Northwestern Medicine study of CSF has discovered its role in cognitive impairment, such as Alzheimer’s disease. This discovery provides a new clue to the process of neurodegeneration, said study lead author David Gate, assistant professor of neurology at Northwestern University Feinberg School of Medicine.
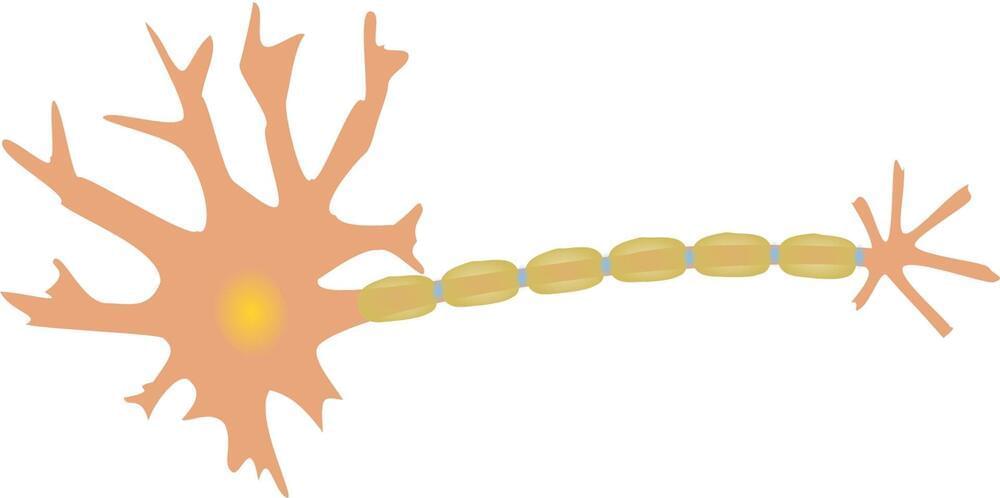
A new study by Burke Neurological Institute (BNI), Weill Cornell Medicine, finds that activation of MAP2K signaling by genetic engineering or non-invasive repetitive transcranial magnetic stimulation (rTMS) promotes corticospinal tract (CST) axon sprouting and functional regeneration after spinal cord injury (SCI) in mice.
RTMS is a noninvasive technique that evokes an electrical field in brain tissue via electromagnetic induction. While an increasing body of evidence suggests that rTMS applied over motor cortex may be beneficial for functional recovery in SCI patients, the molecular and cellular mechanisms that underlie rTMS’ beneficial effects remains unclear.
A new study published in Science Translation Medicine showed that high-frequency rTMS (HF-rTMS) activated MAP2K signaling and enhanced axonal regeneration and functional recovery, suggesting that rTMS might be a valuable treatment option for SCI individuals.