MagicBot, China’s first humanoid robot achieving multi-robot collaboration, enabling flexible production and efficiently handling complex industrial scenarios.



The leading cause of death due to injuries in war is excessive bleeding. A KAIST research team, in which an Army Major participated, has tackled this issue head-on. By developing a next-generation powder-type hemostatic agent that stops bleeding just by spraying it, they have presented an innovative technology that will change the paradigm of combatant survivability.
A joint research team led by Professor Steve Park from the Department of Materials Science and Engineering and Professor Sangyong Jon from the Department of Biological Sciences has developed a powder-type hemostatic agent that forms a powerful hydrogel barrier within approximately one second when sprayed on a wound.
The research was published in Advanced Functional Materials.


An intertwined history: the earth observer and EOS

Our universe is filled with galaxies, in all directions as far as our instruments can see. Some researchers estimate that there are as many as 2 trillion galaxies in the observable universe. At first glance, these galaxies might appear to be randomly scattered across space, but they’re not. Careful mapping has shown that they are distributed across the surfaces of giant cosmic “bubbles” up to several hundred million light-years across. Inside these bubbles, few galaxies are found, so those regions are called cosmic voids. NASA’s Nancy Grace Roman Space Telescope will allow us to measure these voids with new precision, which can tell us about the history of the universe’s expansion.
“Roman’s ability to observe wide areas of the sky to great depths, spotting an abundance of faint and distant galaxies, will revolutionize the study of cosmic voids,” said Giovanni Verza of the Flatiron Institute and New York University, lead author on a paper published in The Astrophysical Journal.
Cosmic recipe The cosmos is made of three key components: normal matter, dark matter, and dark energy. The gravity of normal and dark matter tries to slow the expansion of the universe, while dark energy opposes gravity to speed up the universe’s expansion. The nature of both dark matter and dark energy is currently unknown. Scientists are trying to understand them by studying their effects on things we can observe, such as the distribution of galaxies across space.

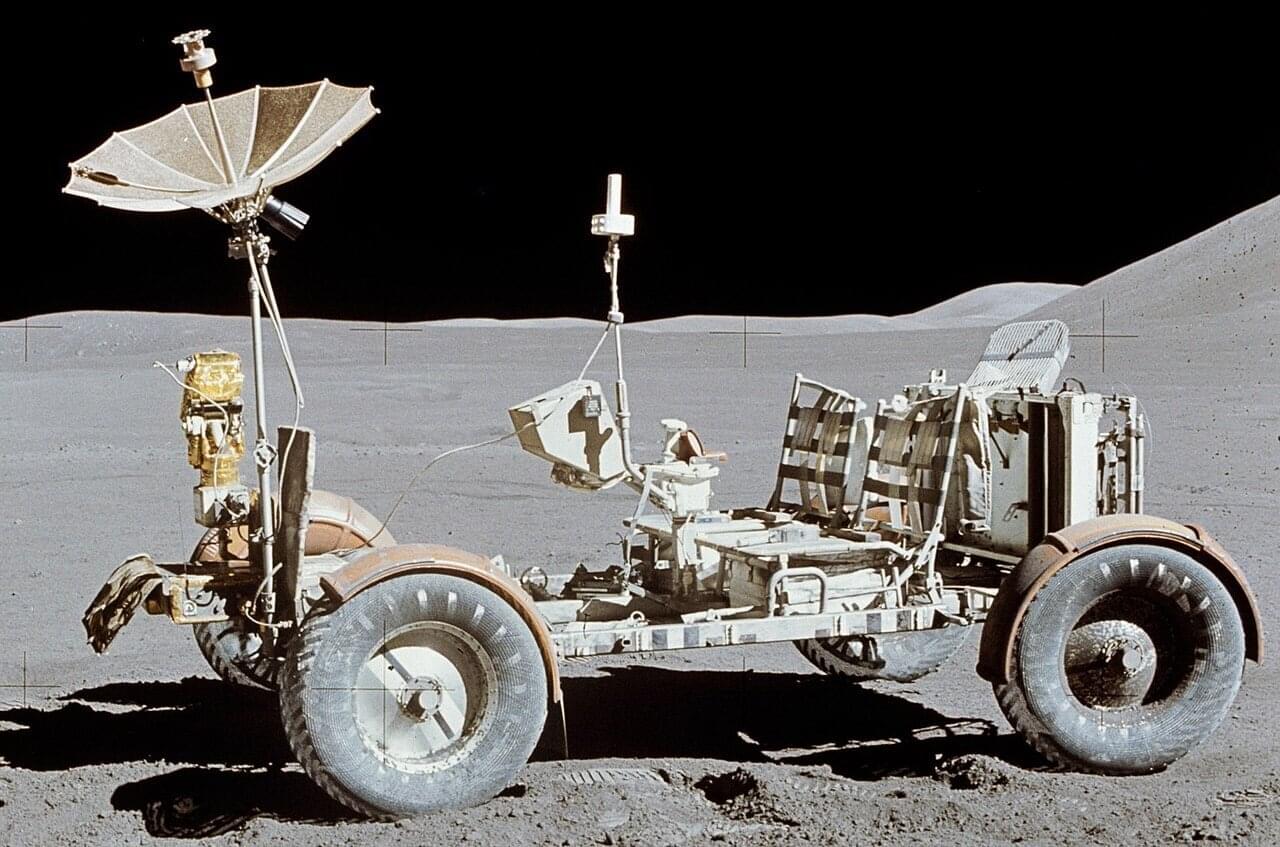
Beneath the moon’s cratered surface lie networks of lava tubes and deep pits, natural caves that could shelter future lunar bases from cosmic radiation and wild temperature swings. These underground structures represent some of the most scientifically valuable areas in the solar system, but they come with the very real challenge of simply getting there.
The entrances to these caves feature steep, rugged terrain with rocks and loose regolith. Small rovers, preferred for lunar exploration because you can deploy many of them to reduce mission risk, face an inherent limitation. Their compact wheels simply can’t climb over obstacles much larger than the wheel diameter itself. Send a swarm of small rovers and even if some fail, others continue the mission. Send one large rover and a single failure ends everything.
Variable diameter wheels are a new thing in lunar exploration and could solve this, expanding when needed to overcome obstacles, then contracting for efficient transport. But building such a wheel for the moon has proven nearly impossible. The lunar environment is uniquely hostile to mechanical systems. Fine, abrasive dust infiltrates everything, and in the airless vacuum, exposed metal surfaces stick together through a process called cold welding. Traditional hinges and joints don’t last long under these conditions.

The treatment was unusual in that alongside talk therapy, May underwent several sessions in a sensory-deprivation chamber: a dark, soundproof room where she floated in a shallow pool of water heated to match the temperature of her skin and saturated with Epsom salts to make her more buoyant. The goal was to blunt May’s external senses, enabling her to feel from within—focusing on the steady thudding of her heart, the gentle flow of air in and out of her lungs, and other internal bodily signals.
The ability to connect with the body’s inner signals is called interoception. Some people are better at it than others, and one’s aptitude for it may change. Life events can also bolster or damage a person’s interoceptive skills. Sahib Khalsa, a psychiatrist and neuroscientist at the University of California, Los Angeles, and his colleagues think a disrupted interoception system might be one of the driving forces behind anorexia nervosa. So they decided to repurpose a decades-old therapy called flotation-REST (for “reduced environmental stimulation therapy”) and launched a trial with it in 2018. They. hypothesized that in people with anorexia and some other disorders, an underreliance on internal signals may lead to an overreliance on external ones, such as how one looks in the mirror, that ultimately causes distorted body image, one of the key factors underlying these conditions. “When they’re in the float environment, they experience internal signals more strongly,” Khalsa says. “And having that experience may then confer a different understanding of the brain-body relationship that they have.”
Disruptions in interoception may underlie anxiety, eating disorders, and other mental health ailments.
By Diana Kwon edited by Jeanna Bryner.