Recorded 10 February 2026. Sebastien Bubeck of OpenAI presents “A Combinatorics Problem” at IPAM’s AI for Science Kickoff. Learn more online at: https://www.ipam.ucla.edu/programs/sp… AI for Science Kickoff 2026: This inaugural event brings together the pioneers who are defining how AI will accelerate scientific discovery — from Nobel and Fields Medal laureates to the leaders shaping AI innovation across academia, research labs, and industry. The event features keynote talks by leading AI Scientists and Mathematicians, as well as panel discussions focusing on perspectives on AI from three sides: Mathematics, Higher Education, and Industry. This event is organized jointly by IPAM, the UCLA Division of Physical Sciences, the SAIR Foundation and the World Leading Scientists Institute.
Get the latest international news and world events from around the world.

Helion hits new fusion milestone: D-T fusion and 150M°C plasma temperatures
Helion has achieved a significant milestone in fusion energy by successfully demonstrating deuterium-tritium fusion with plasma temperatures reaching 150 million degrees Celsius.
## Questions to inspire discussion.
Fusion Performance Achievements.
🔥 Q: What fusion performance records did Helion’s Polaris achieve?
A: Polaris became the first privately funded fusion machine to demonstrate measurable deuterium-tritium (DT) fusion while reaching plasma temperatures exceeding 150 million degrees Celsius, proving the ability to compress and hold fusion plasma for more pressure, more heat, and more fusion.
Operational Execution.

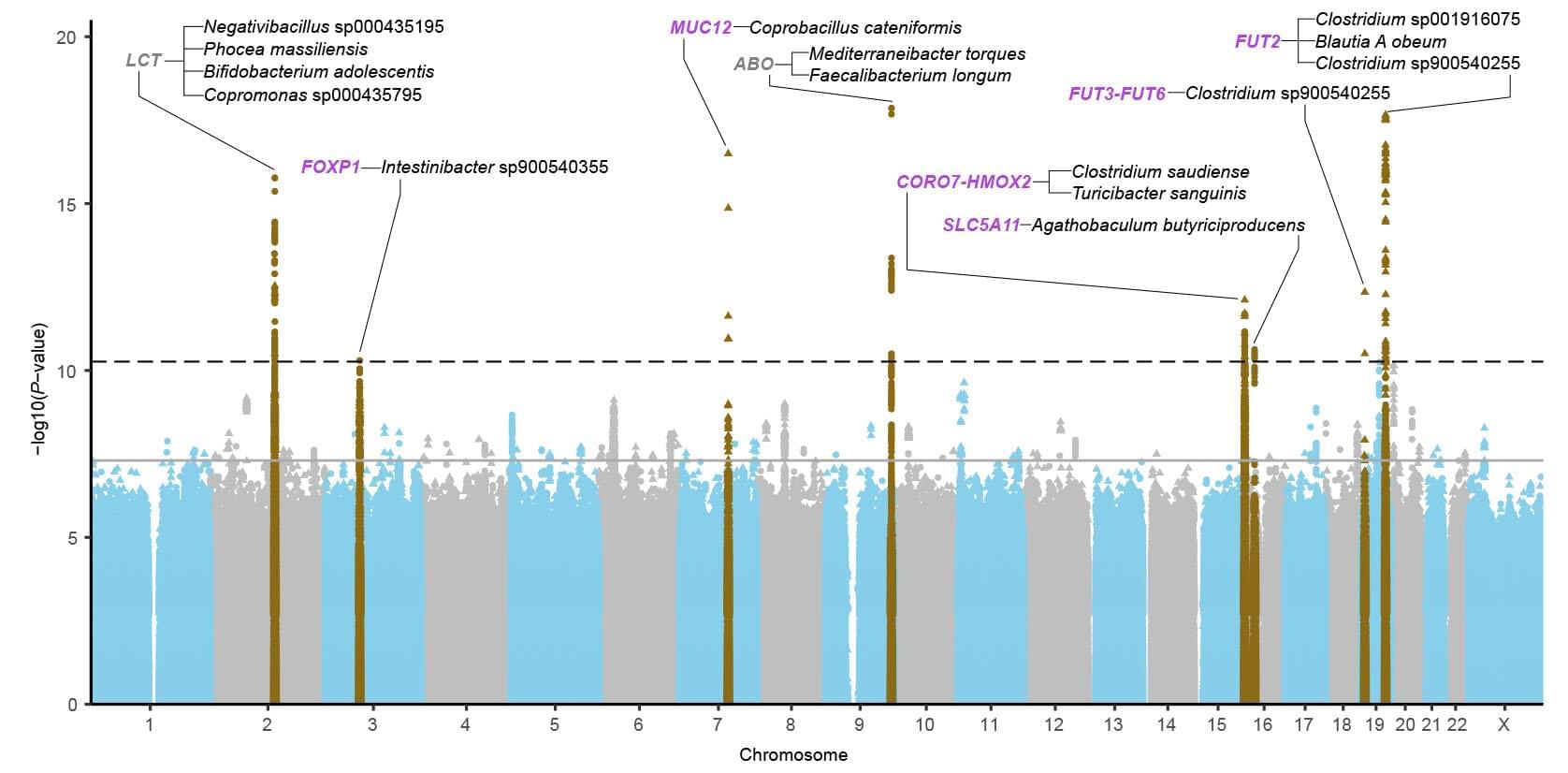
Studies show 11 genetic variants affect gut microbiome
In two new studies on 28,000 individuals, researchers are able to show that genetic variants in 11 regions of the human genome have a clear influence on which bacteria are in the gut and what they do there. Only two genetic regions were previously known. Some of the new genetic variants can be linked to an increased risk of gluten intolerance, hemorrhoids and cardiovascular diseases.
The studies are published in the journal Nature Genetics.
The community of bacteria living in our gut, or gut microbiome, has become a hot research area in recent years because of its great significance for health and disease. However, the extent to which our genes determine which bacteria are present in the intestines has been unclear. Until now, it has only been possible to link a few genetic variants to the composition of the gut microbiome with certainty.

Antarctica sits above Earth’s strongest ‘gravity hole.’ Now we know how it got that way
Gravity feels reliable—stable and consistent enough to count on. But reality is far stranger than our intuition. In truth, the strength of gravity varies over Earth’s surface. And it is weakest beneath the frozen continent of Antarctica after accounting for Earth’s rotation.
A new study reveals how achingly slow rock movements deep under Earth’s surface over tens of millions of years led to today’s Antarctic gravity hole. The study highlights that the timing of changes in the Antarctic gravity low overlaps with major changes in Antarctica’s climate, and future research could reveal how the shifting gravity might have encouraged the growth of the frozen continent’s climate-defining ice sheets.
“If we can better understand how Earth’s interior shapes gravity and sea levels, we gain insight into factors that may matter for the growth and stability of large ice sheets,” said Alessandro Forte, Ph.D., a professor of geophysics at the University of Florida and co-author of the new study recreating the Antarctic gravity hole’s past.

Brain mechanism behind ‘flashes of intuition’
Despite decades of research, the mechanisms behind fast flashes of insight that change how a person perceives their world, termed “one-shot learning,” have remained unknown. A mysterious type of one-shot learning is perceptual learning, in which seeing something once dramatically alters our ability to recognize it again.
Now a new study, the researchers address the moments when we first recognize a blurry object, a primal ability that enabled our ancestors to avoid threats.
Published in Nature Communications, the new work pinpoints for the first time the brain region called the high-level visual cortex (HLVC) as the place where “priors” — images seen in the past and stored — are accessed to enable one-shot perceptual learning.
“Our work revealed, not just where priors are stored, but also the brain computations involved,” said co-senior study author.
Importantly, past studies had shown that patients with schizophrenia and Parkinson’s disease have abnormal one-shot learning, such that previously stored priors overwhelm what a person is presently looking at to generate hallucinations.
“This study yielded a directly testable theory on how priors act up during hallucinations, and we are now investigating the related brain mechanisms in patients with neurological disorders to reveal what goes wrong,” added the author.
The research team is also looking into likely connections between the brain mechanisms behind visual perception and the better-known type of “aha moment” when we comprehend a new idea. ScienceMission sciencenewshighlights.


Discussing the implication of DNA methylation in human diseases
DNA methylation plays a critical role in gene expression regulation and has emerged as a robust biomarker of biological age. This modification will become heavier or site drift along with aging. Recently, it is termed epigenetic clocks—such as Horvath, Hannum, PhenoAge, and GrimAge—leverage specific methylation patterns to accurately predict age-related decline, disease risk, and mortality. These tools are now widely applied across diverse tissues, populations, and disease contexts. Beyond age-related loss of methylation control, accelerated DNA methylation age has been linked to environmental exposures, lifestyle factors, and chronic diseases, further reinforcing its value as a dynamic and clinically relevant marker of biological aging. DNA methylation is reshaping our understanding of aging and disease risk, with promising implications for preventive medicine and interventions aimed at promoting healthy longevity. However, it must be admitted that some challenges remain, including limited generalizability across populations, an unclear mechanism, and inconsistent longitudinal performance. In this review, we examine the biological foundations of DNA methylation, major advances in epigenetic clock development, and their expanding applications in aging research, disease prediction and health monitoring.
Aging is a complex, multifactorial process that affects nearly all biological systems. While chronological age simply measures the passage of time from birth, biological age reflects the functional state and health of an individual’s tissues and organs (Kiselev et al., 2025). This distinction is critical, as individuals of the same chronological age often exhibit markedly different biological conditions, disease risks, and mortality trajectories (Dugue et al., 2018). Therefore, biological age potentially serves as a more meaningful measure of aging-related decline and is increasingly used to assess overall health status, predict disease onset, and evaluate the effectiveness of interventions aimed at promoting healthy longevity (Dugue et al., 2018; Petkovich et al., 2017).
Among various biomarkers proposed to estimate biological age, epigenetic modifications—particularly DNA methylation—have emerged as one of the most reliable and informative (Dugue et al., 2018). In epigenetics, DNA methylation involves the addition of a methyl group to the 5′ position of cytosine residues, typically at CpG dinucleotides, which can regulate gene expression without altering the underlying DNA sequence. Moreover, DNA methylation can be accurately measured by sequencing at methylated sites with bisulfate treatment (Zhang et al., 2012). Age-related changes in DNA methylation pattern are not random; they occur at specific genomic locations. These methylated sites are picked and constitute come patterns, by which scientists can construct “epigenetic clocks” to precisely estimate a person’s biological age based on their DNA modification. As people grow older, their methylation profiles shift in predictable ways (Kiselev et al., 2025; Horvath, 2013; Horvath and Raj, 2018).
