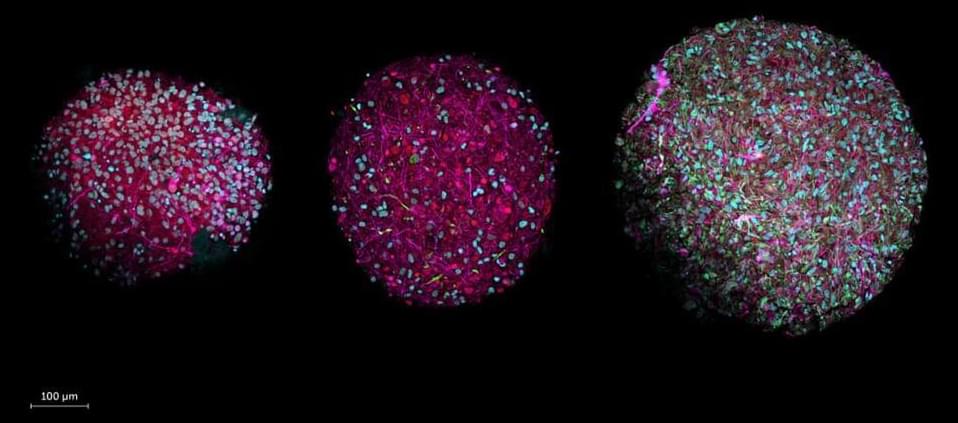
Despite AI’s impressive track record, its computational power pales in comparison with that of the human brain. Scientists today unveil a revolutionary path to drive computing forward: organoid intelligence (OI), where lab-grown brain organoids serve as biological hardware. “This new field of biocomputing promises unprecedented advances in computing speed, processing power, data efficiency, and storage capabilities – all with lower energy needs,” say the authors in an article published in Frontiers in Science.
Artificial intelligence (AI) has long been inspired by the human brain. This approach proved highly successful: AI boasts impressive achievements – from diagnosing medical conditions to composing poetry. Still, the original model continues to outperform machines in many ways. This is why, for example, we can ‘prove our humanity’ with trivial image tests online. What if instead of trying to make AI more brain-like, we went straight to the source?
Scientists across multiple disciplines are working to create revolutionary biocomputers where three-dimensional cultures of brain cells, called brain organoids, serve as biological hardware. They describe their roadmap for realizing this vision in the journal Frontiers in Science.