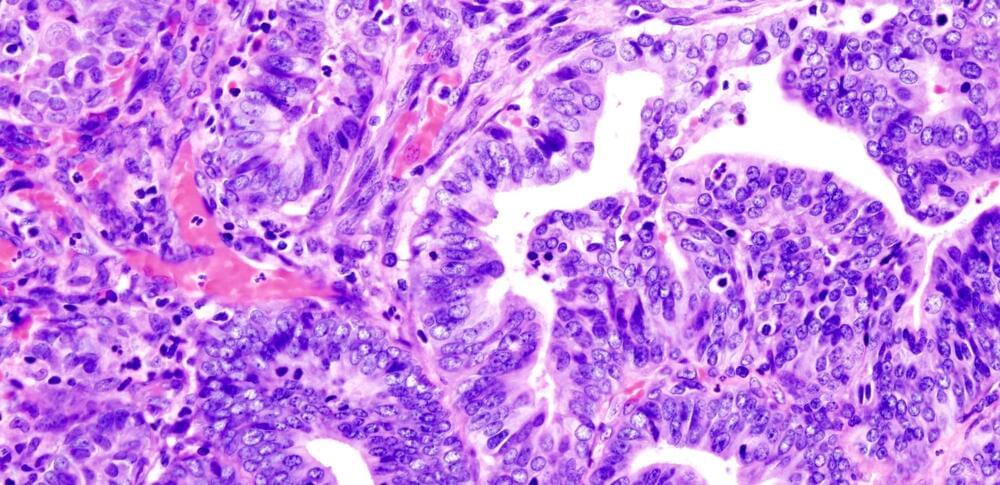
Detailed molecular characterization of endometrial carcinomas identifies potential markers for guiding more precise treatment.


For his work on techniques to generate quantum dots of uniform size and color, Bawendi is honored along with Louis Brus and Alexei Ekimov.
Moungi Bawendi, the Lester Wolfe Professor of Chemistry at MIT and a leader in the development of tiny particles known as quantum dots, has won the Nobel Prize in Chemistry for 2023. He will share the prize with Louis Brus of Columbia University and Alexei Ekimov of Nanocrystals Technology, Inc.
The researchers were honored for their work in discovering and synthesizing quantum dots — tiny particles of matter that emit exceptionally pure light. In its announcement this morning, the Nobel Foundation cited Bawendi for work that “revolutionized the chemical production of quantum dots, resulting in almost perfect particles.”

Putting a greater emphasis on the development of AGI — artificial general intelligence. CEO Sam Altman has described AGI as “the equivalent of a median human that you could hire as a co-worker.”
The AI startup says its singular goal is to build “safe, beneficial” artificial general intelligence, noting that anything else is “out of scope.”

Where reliability matters, as it does in energy, resilience against cyberattacks enhances a company’s reputation. Disruptions damage that reputation.
In 2021, a ransomware attack shut down Colonial Pipeline operations for six days. Gas shortages in the eastern US, economic turmoil, and eye-catching headlines resulted. Interest in cybersecurity for critical infrastructure intensified — and many leaders seemed to learn the wrong lesson.
Energy sector leaders often take cyber vulnerabilities seriously only after a significant breach. Experiencing a loss (or watching someone else’s) makes companies tighten cybersecurity to avoid similar losses. This pattern emphasizes the loss-avoidance aspects of cybersecurity. Yet thinking of cybersecurity solely as loss avoidance misses a key value generator cybersecurity provides: trust.
Companies that get cybersecurity right earn trust. That trust matters in two ways: It supports brand or company reputation, and it allows for forward innovation.


Salk Institute researchers, as part of a larger collaboration with research teams around the world, analyzed more than half a million brain cells from three human brains to assemble an atlas of hundreds of cell types that make up a human brain in unprecedented detail.
The research, published in a special issue of the journal Science on October 13, 2023, is the first time that techniques to identify brain cell subtypes originally developed and applied in mice have been applied to human brains.
“These papers represent the first tests of whether these approaches can work in human brain samples, and we were excited at just how well they translated,” says Professor Joseph Ecker, director of Salk’s Genomic Analysis Laboratory and a Howard Hughes Medical Institute investigator. “This is really the beginning of a new era in brain science, where we will be able to better understand how brains develop, age, and are affected by disease.”
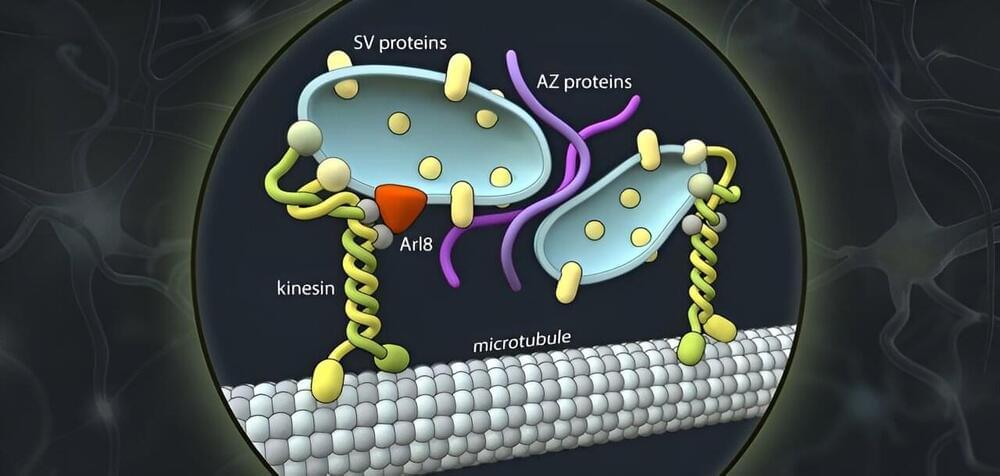
Whether in the brain or in the muscles, wherever there are nerve cells, there are synapses. These contact points between neurons form the basis for the transmission of excitation, the communication between neurons. As in any communication process, there is a sender and a receiver: Nerve cell processes called axons generate and transmit electrical signals thereby acting as signal senders.
Synapses are points of contact between axonal nerve terminals (the pre-synapse) and post-synaptic neurons. At these synapses, the electrical impulse is converted into chemical messengers that are received and sensed by the post-synapses of the neighboring neuron. The messengers are released from special membrane sacs called synaptic vesicles.
As well as transmitting information, synapses can also store information. While the structure and function of synapses are comparably well understood, little is known about how they are formed.
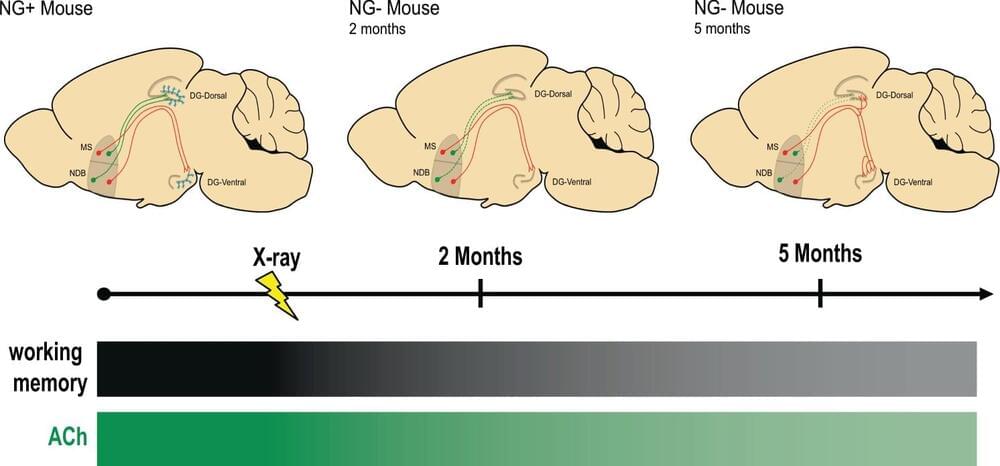
A quarter-century ago, researchers discovered that adults, not just developing infants, can generate new brain cells, a process called neurogenesis. But it’s still not clear what role these new neurons play in health or disease.
In a new mouse study, Columbia University researchers found that neurogenesis in adults is critical for maintaining brain circuits that support working memory across the lifespan and chronic loss of adult neurogenesis causes progressive memory loss, like that seen in age-related cognitive decline and Alzheimer’s disease in humans.
The study, “Adult-born neurons maintain hippocampal cholinergic inputs and support working memory during aging,” was published in July in the journal Molecular Psychiatry.

A study by researchers in the laboratory of Dr. Huda Zoghbi, distinguished service professor at Baylor College of Medicine and director of the Jan and Dan Duncan Neurological Research Institute (Duncan NRI) at Texas Children’s Hospital, has discovered that diminished memory recall in Rett syndrome mice can be restored by activating specific inhibitory cells in the hippocampus. The findings are published in the current edition of Neuron.
Rett syndrome is a neurodevelopmental disorder characterized by loss of acquired cognitive, motor, language and social skills after the first year of life as well as profound learning and memory impairments. In particular, contextual memories, those that encode an event and the circumstances in which the event was experienced, are diminished in mouse models of Rett syndrome. Previous research has suggested that diminished contextual memories result from disruptions in the finely tuned balance between excitatory and inhibitory synaptic inputs that constantly bombard hippocampal neurons.
Zoghbi’s team hypothesized that disruptions in this balance may alter the size and composition of ensembles of hippocampal neurons needed to encode a contextual memory. Using a miniature microscope, they directly monitored these ensembles as mice recalled a fearful experience. They found that Rett mice have larger and more correlated ensembles of neurons than wild-type mice, suggesting that hippocampal pyramidal neurons are not receiving enough inhibition in Rett mice.
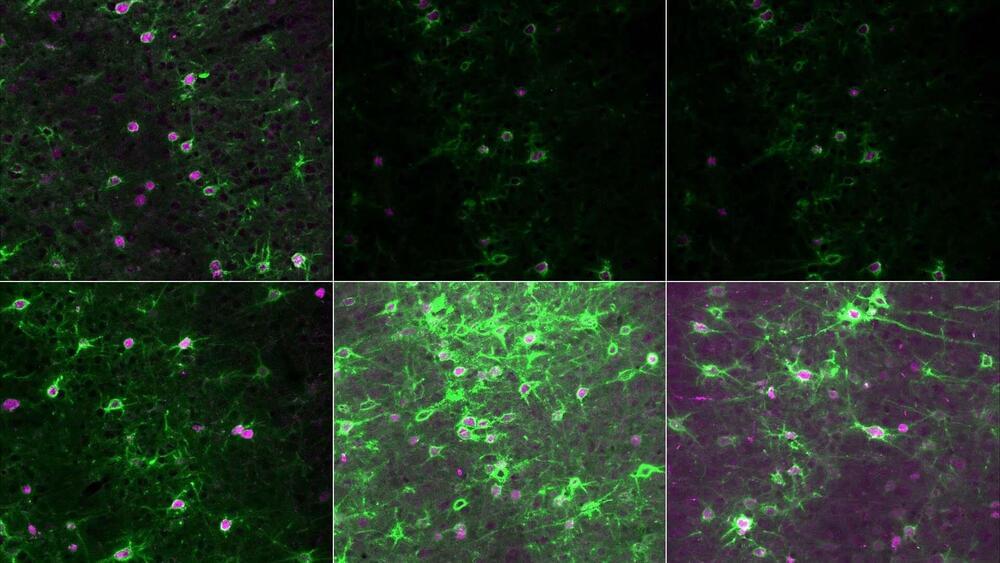
Developing brains become shaped by the sights, sounds, and experiences of early life. The brain’s circuits grow more stable as we age. However, some experiences later in life open up opportunities for these circuits to be rapidly rewired. New research from Cold Spring Harbor Laboratory Associate Professor Stephen Shea helps explain how the brain adapts during a critical period of adulthood: the time when new mothers learn to care for their young.
Shea’s work in mice shows how this learning process is disrupted when a small set of neurons lack a protein called MECP2. In humans, MECP2 dysfunction causes the rare neurodevelopmental disorder Rett syndrome. Shea’s findings could point researchers toward the brain circuits involved in Rett syndrome and potential treatment strategies. His research could also have implications for more common neurological conditions.
Shea explains, “It’s not lost on us that Rett syndrome patients have difficulty interpreting and producing language. Difficulties with communicating are widespread in autism spectrum disorders. One of the reasons we study Rett syndrome is that this may be a valuable model for other forms of autism.”