UTA research could help restore proper cholesterol levels, stopping many diseases in their tracks

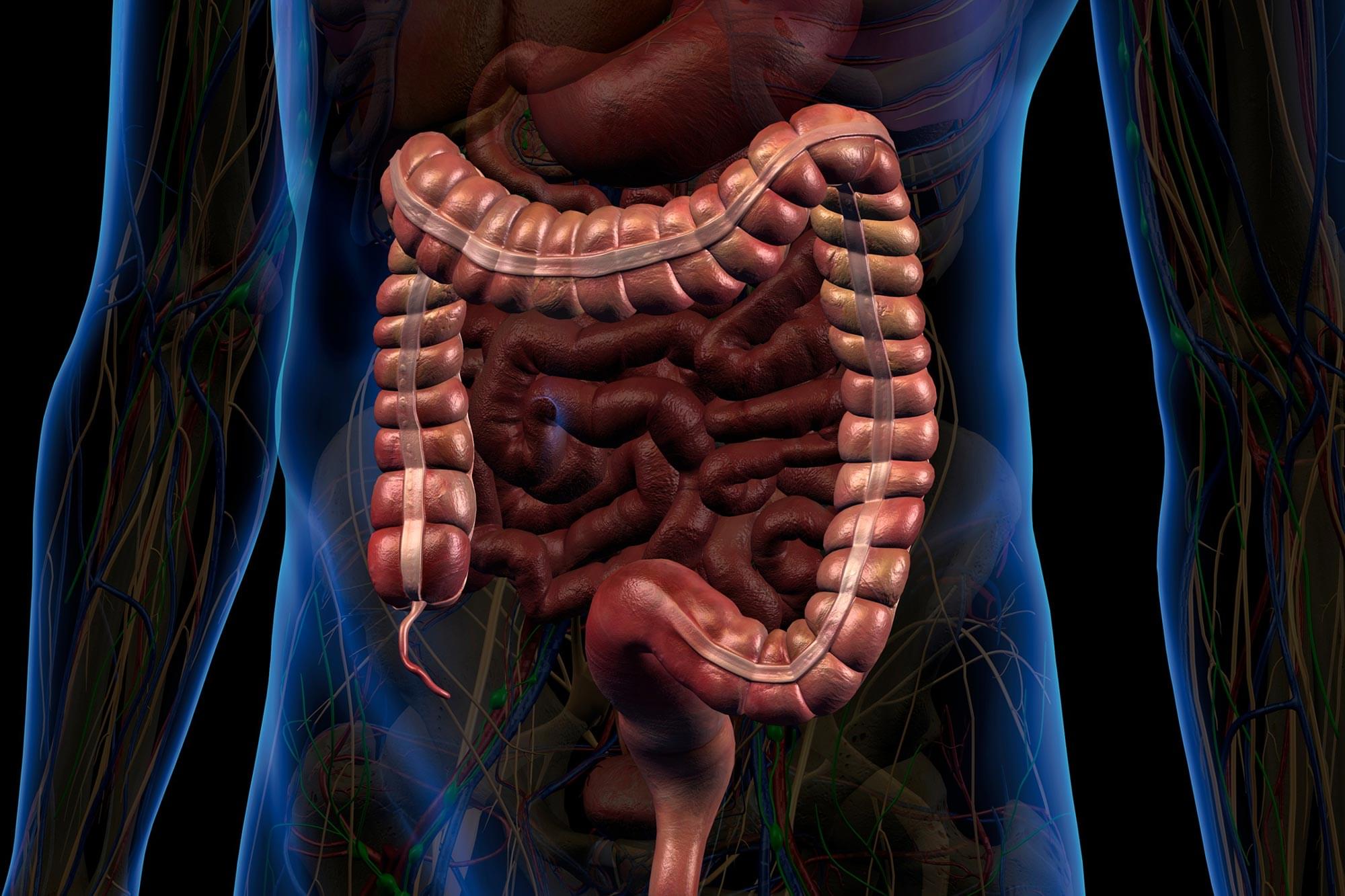
Researchers are gaining a deeper appreciation for the critical role the gastrointestinal (GI) tract plays in maintaining overall health. Beyond its primary responsibilities in digestion, the GI system contributes to the production of hormones, immune cells, and neurotransmitters that influence brain function and emotional well-being.
Because of this, the GI tract contains a wide array of biomarkers that are valuable for diagnosing, tracking, and managing disease—from short-chain fatty acids associated with metabolic syndrome to cytokines linked to inflammation.
However, current technologies fall short when it comes to capturing this biochemical information directly from the GI tract. Existing methods, such as fecal sampling and tissue biopsies, are often invasive, costly, and unable to deliver continuous or comprehensive real-time data throughout the length of the digestive system.
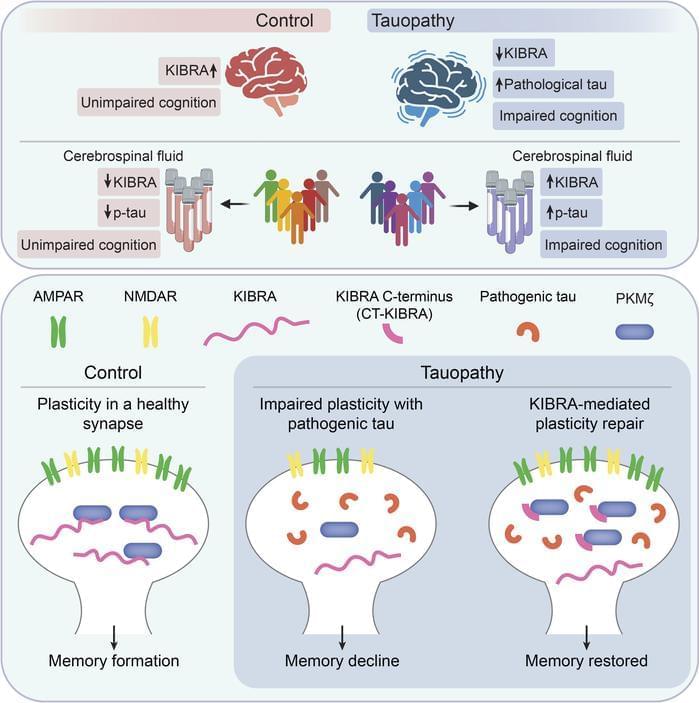
Representing a key mechanism that underlies memory loss in Alzheimer’s disease (AD) and related tauopathies. Here, we found that reduced levels of the memory-associated protein KIdney/BRAin (KIBRA) in the brain and increased KIBRA protein levels in cerebrospinal fluid are associated with cognitive impairment and pathological tau levels in disease. We next defined a mechanism for plasticity repair in vulnerable neurons using the C-terminus of the KIBRA protein (CT-KIBRA). We showed that CT-KIBRA restored plasticity and memory in transgenic mice expressing pathogenic human tau; however, CT-KIBRA did not alter tau levels or prevent tau-induced synapse loss. Instead, we found that CT-KIBRA stabilized the protein kinase Mζ (PKMζ) to maintain synaptic plasticity and memory despite tau-mediated pathogenesis. Thus, our results distinguished KIBRA both as a biomarker of synapse dysfunction and as the foundation for a synapse repair mechanism to reverse cognitive impairment in tauopathy.
1Buck Institute for Research on Aging, Novato, California, USA.
2Memory and Aging Center, Department of Neurology, University of California San Francisco, San Francisco, California, USA.
3Gladstone Institutes, San Francisco, Califoria, USA.
4Weill Institute for Neurosciences, Department of Pathology, University of California San Francisco, San Francisco, California, USA.

Using an inexpensive electrode coated with DNA, MIT researchers have designed disposable diagnostics that could be adapted to detect a variety of diseases, including cancer or infectious diseases such as influenza and HIV.
These electrochemical sensors make use of a DNA-chopping enzyme found in the CRISPR gene-editing system. When a target such as a cancerous gene is detected by the enzyme, it begins shearing DNA from the electrode nonspecifically, like a lawnmower cutting grass, altering the electrical signal produced.
One of the main limitations of this type of sensing technology is that the DNA that coats the electrode breaks down quickly, so the sensors can’t be stored for very long and their storage conditions must be tightly controlled, limiting where they can be used. In a new study, MIT researchers stabilized the DNA with a polymer coating, allowing the sensors to be stored for up to two months, even at high temperatures. After storage, the sensors were able to detect a prostate cancer gene that is often used to diagnose the disease.
In recent years, some scientists and advocates have warned that playing contact sports like football and hockey may increase the risk of brain diseases like Alzheimer’s disease or chronic traumatic encephalopathy (CTE) due to a buildup of a specific protein in the brain.
But a new Northwestern Medicine study of 174 donated brains, including some from former high school and college football players, pumps the brakes on that theory.
“The long and short of it is no, this protein in this specific brain region is not increased in people who played football at the amateur level. It throws a little bit of cold water on the current CTE narrative,” said corresponding author Dr. Rudolph Castellani, professor of pathology at Northwestern University Feinberg School of Medicine and a Northwestern Medicine neuropathologist.

Australian scientists, including at the Charles Perkins Centre, University of Sydney, have successfully developed a research system that uses ‘biological artificial intelligence’ to design and evolve molecules with new or improved functions directly in mammal cells. The researchers said this system provides a powerful new tool that will help scientists develop more specific and effective research tools or gene therapies.
Named PROTEUS (PROTein Evolution Using Selection) the system harnesses ‘directed evolution’, a lab technique that mimics the natural power of evolution. However, rather than taking years or decades, this method accelerates cycles of evolution and natural selection, allowing them to create molecules with new functions in weeks.
This could have a direct impact on finding new, more effective medicines. For example, this system can be applied to improve gene editing technology like CRISPR to improve its effectiveness.


Imagine having a conversation where every gesture and glance feels like a test. You’re juggling eye contact, facial expressions, and tone of voice, all while trying to keep up with the words. You might miss something, or someone might misread you.
In a new study, published in PLOS One, autistic adults describe the intense mental effort it takes to navigate nonverbal communication (NVC).
Researchers reviewed 362 firsthand accounts on the online forum WrongPlanet.net, where autistic adults openly talk about communication challenges. They focused on posts about nonverbal communication—like eye contact, tone of voice, gestures, and facial expressions—and reviewed 26 discussion threads to better understand from autistic adults what it is like to communicate in daily life.
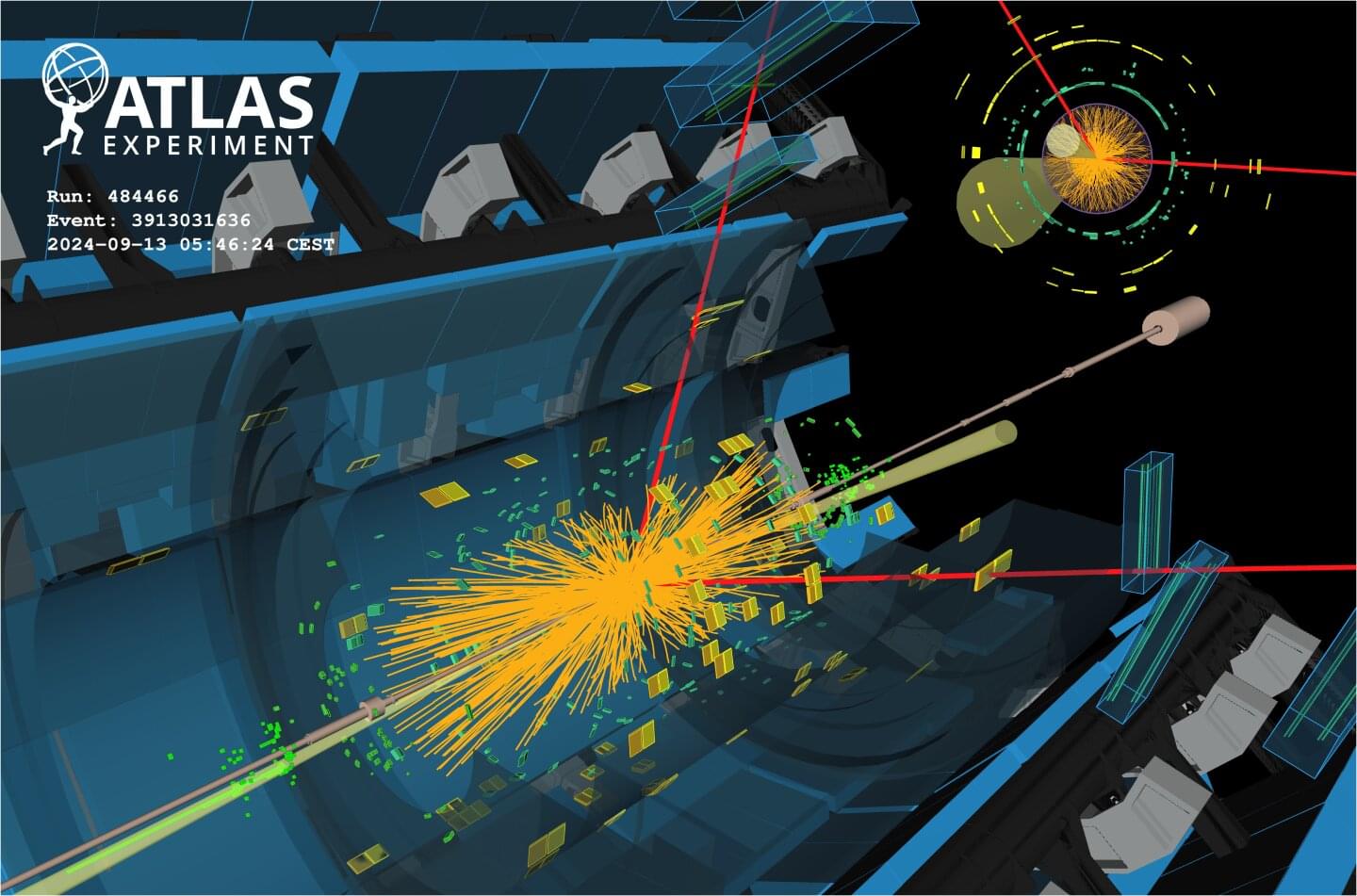
The ATLAS collaboration finds evidence of Higgs-boson decays to muons and improves sensitivity to Higgs-boson decays to a Z boson and a photon.
Studies of the properties of the Higgs boson featured prominently in the program of the major annual physics conference, the 2025 European Physical Society Conference on High Energy Physics (EPS-HEP), held this week in Marseille, France. Among the results presented by the ATLAS collaboration were two results narrowing in on two exceptionally rare Higgs-boson decays.
The first process under study was the Higgs-boson decay into a pair of muons (H→μμ). Despite its scarceness—occurring in just 1 out of every 5,000 Higgs decays—this process provides the best opportunity to study the Higgs interaction with second-generation fermions and shed light on the origin of mass across different generations. Up to now, the interactions of the Higgs boson with matter particles have only been observed for particles from the third, heaviest, generation: the tau lepton and the top and bottom quarks.