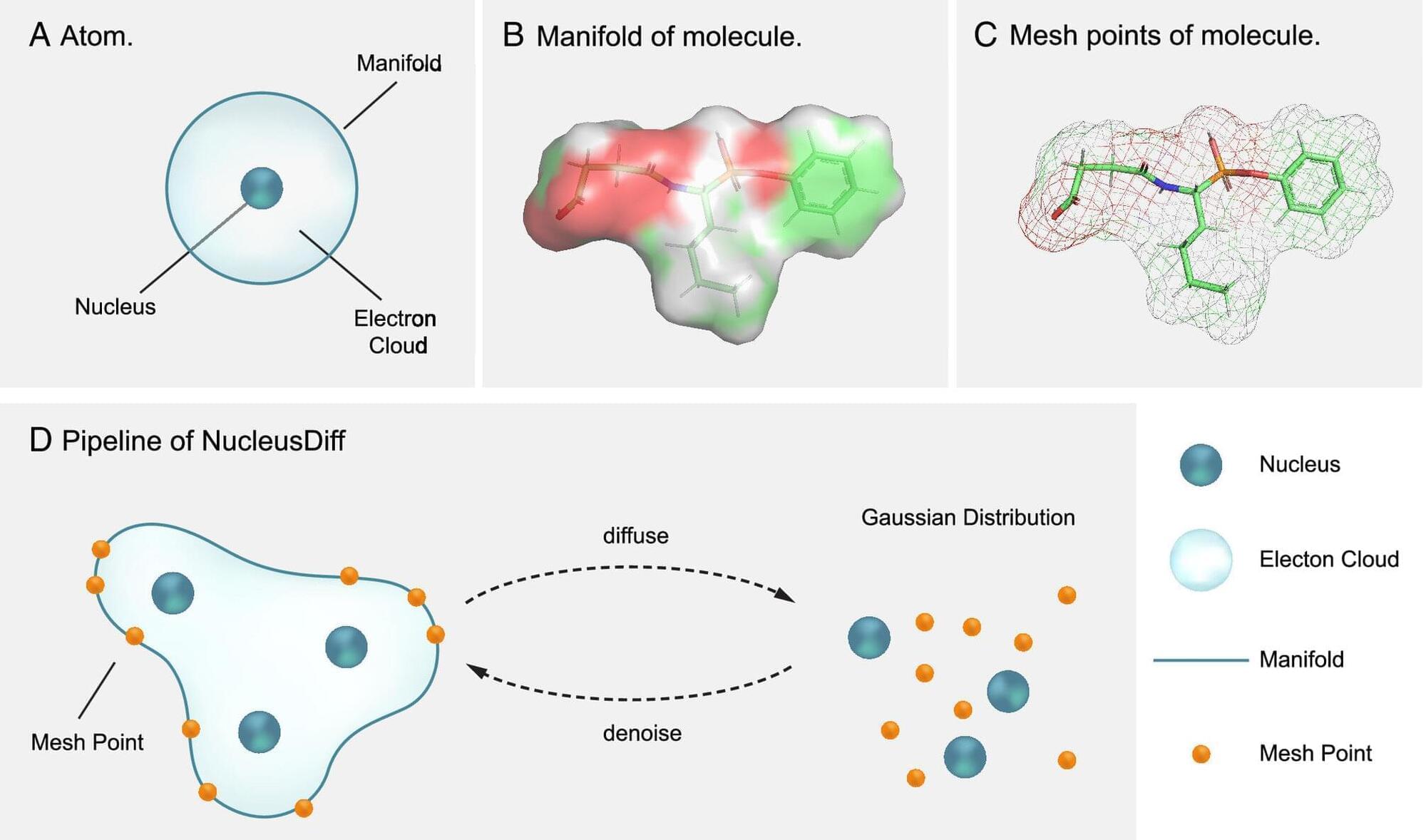
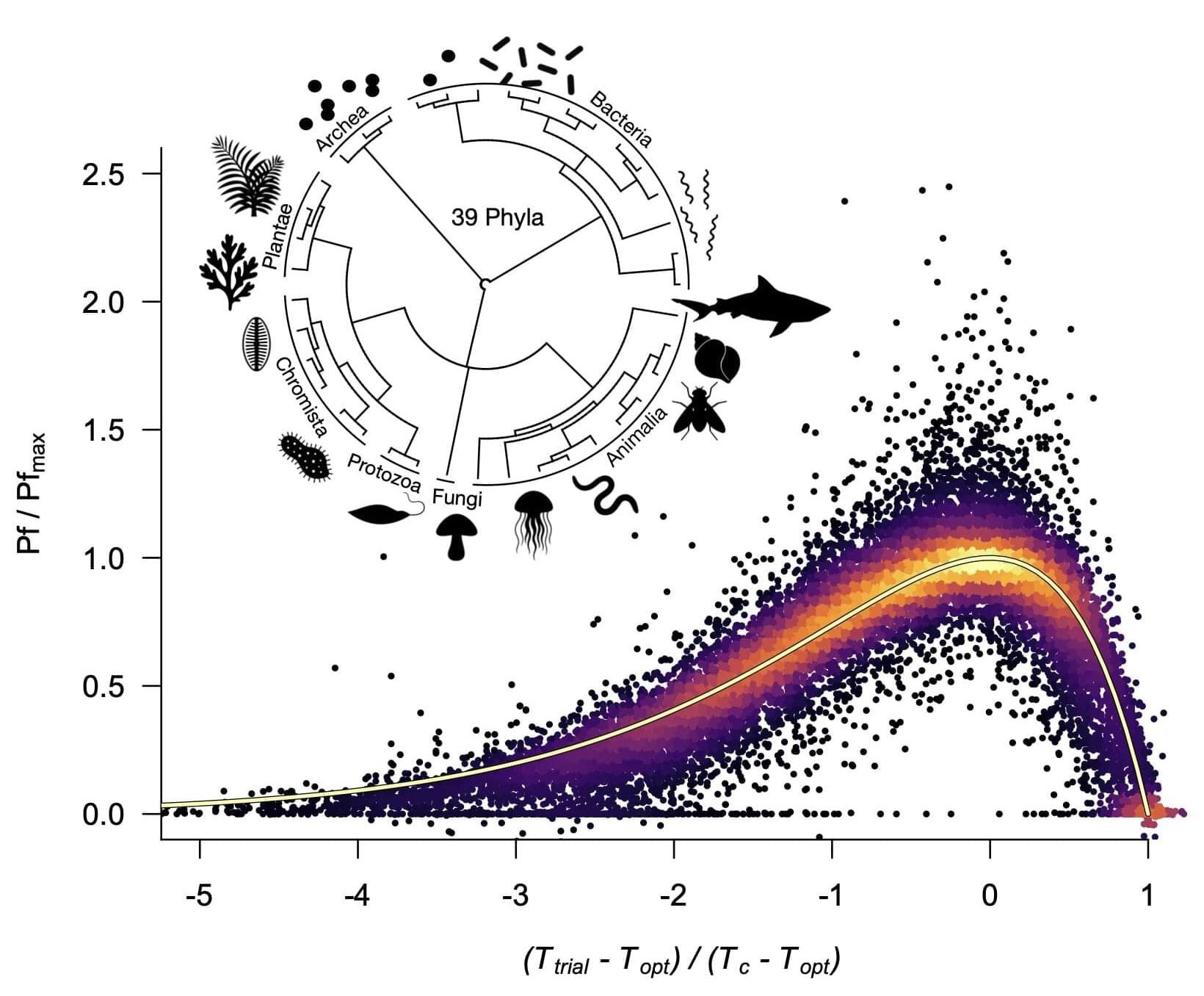
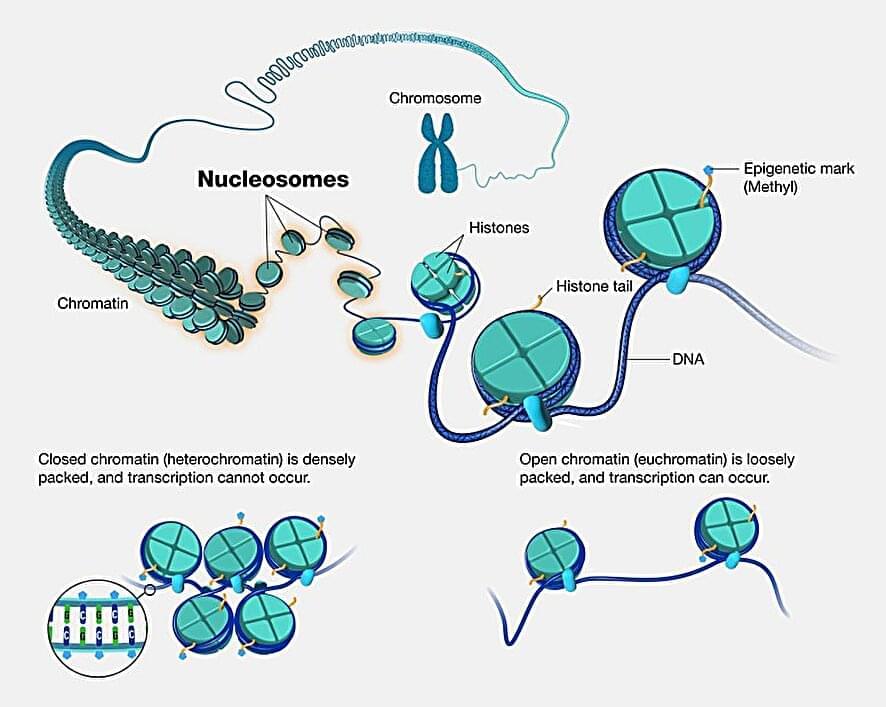
When machine learning is used to suggest new potential scientific insights or directions, algorithms sometimes offer solutions that are not physically sound.
Take, for example, AlphaFold, the AI system that predicts the complex ways in which amino acid chains will fold into 3D protein structures. The system sometimes suggests “unphysical” folds—configurations that are implausible based on the laws of physics —especially when asked to predict the folds for chains that are significantly different from its training data.
To limit this type of unphysical result in the realm of drug design, Anima Anandkumar, Bren Professor of Computing and Mathematical Sciences at Caltech, and her colleagues have introduced a new machine learning model called NucleusDiff, which incorporates a simple physical idea into its training, greatly improving the algorithm’s performance.