Researchers have developed computationally simple robots, called particles, that cluster and form a single “particle robot” that completes tasks.
Get the latest international news and world events from around the world.
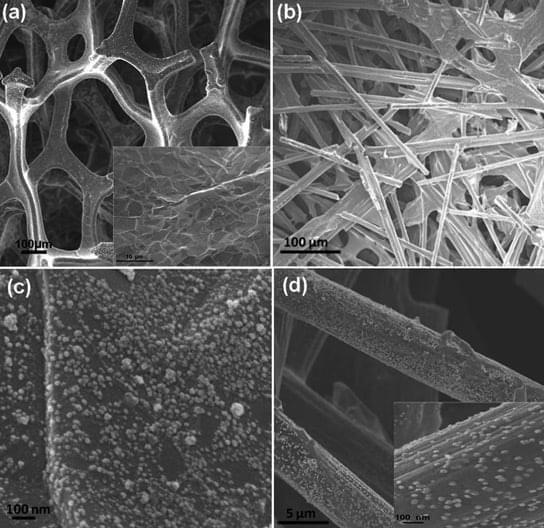
New 3D Form of Graphene May Lead to Flexible Electronics
Graphene can support 50,000 times its own weight and can spring back into shape after being compressed by up to 80%. Graphene also has a much lower density than comparable metal-based materials. A new super-elastic, three-dimensional form of graphene can conduct electricity, and will probably pave t
Ultra-Strong Artificial Muscles Made From Carbon Nanotubes
In order to showcase ultra-strong artificial muscles, Ray Baughman from the University of Texas at Dallas and his colleagues built a catapult.
The scientists published their findings in the journal Science. The device contains yarns similar in diameter to human hair, spun from carbon nanotubes and soaked in paraffin wax. When a current is passed through the yarn, the wax heats up and expands. As the yarn swells, its particular helical weave causes it to shorten, and the muscle contracts. As it cools, the yarn relaxes and returns to its original length. When coiled lightly or heated to high enough temperatures, wax-free yarns behave in the same fashion.
The torque produced by the twisting and untwisting of the yarns is sufficient to power a miniature catapult. The yarn can haul 200 times the weight that a natural muscle of the same size can, and generates more torque than a large electric motor if compared by weight. Currently, the available manufacturing techniques have limited the weight of the yarn. They can make yarn that lifts up 50 grams. That doesn’t sound like much, but researchers have shown the nanotube yarns lifting loads as much as 50,000 times greater than their own weight.



“I Beat Gravity”: American Scientists Stunned as Revolutionary Anti-Gravity Invention Destroys Laws of Physics in Live Demo
IN A NUTSHELL 🚀 A new propulsion technology claims to revolutionize space travel by generating thrust without expelling propellant, challenging established physical laws. 📚 The concept echoes the controversial EmDrive, which failed scientific validation, highlighting the need for rigorous testing of bold claims. 🌟 Charles Buhler and his team, including experts from NASA and Blue


Light fields with extraordinary structure: Plasmonic skyrmion bags
A research group at the University of Stuttgart has manipulated light through its interaction with a metal surface so that it exhibits entirely new properties. The researchers have published their findings in Nature Physics.
“Our results add another chapter to the emerging field of skyrmion research,” proclaims Prof. Harald Giessen, head of the Fourth Physics Institute at the University of Stuttgart, whose group achieved this breakthrough. The team demonstrated the existence of “skyrmion bags” of light on the surface of a metal layer.
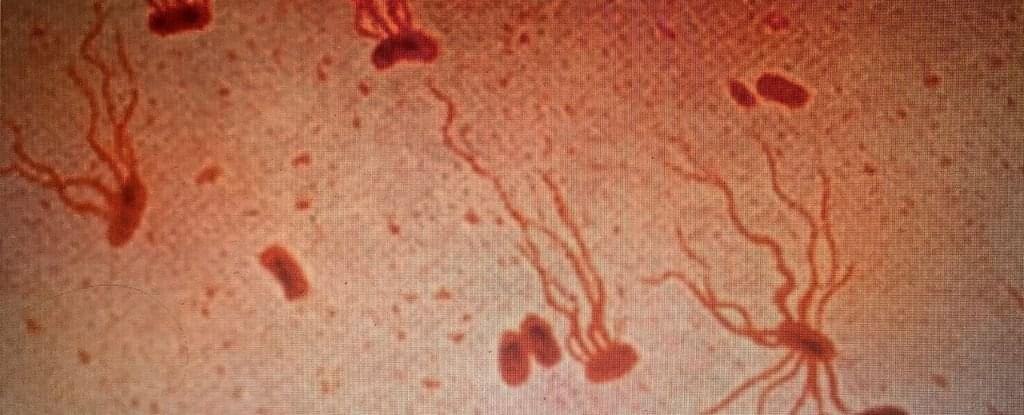
Ancient Killer Is Rapidly Becoming Resistant to Antibiotics, Study Warns
Typhoid fever might be rare in developed countries, but this ancient threat, thought to have been around for millennia, is still very much a danger in our modern world.
According to research published in 2022, the bacterium that causes typhoid fever is evolving extensive drug resistance, and it’s rapidly replacing strains that aren’t resistant.
Currently, antibiotics are the only way to effectively treat typhoid, which is caused by the bacterium Salmonella enterica serovar Typhi (S Typhi). Yet over the past three decades, the bacterium’s resistance to oral antibiotics has been growing and spreading.
Nanobots: Tiny Tech, Infinite Possibilities! #science
Nanobots aren’t just microscopic machines—they could come in countless shapes and sizes, each designed for a unique purpose. From medical nanobots that repair cells to swarming micro-robots that build structures at the atomic level, the future of nanotechnology is limitless. Could these tiny machines revolutionize medicine, industry, and even space exploration? #Nanotech #Nanobots #FutureTech #Science #Innovation …