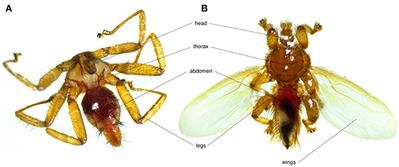
Bats are the second most diverse mammalian group, playing keystone roles in ecosystems but also act as reservoir hosts for numerous pathogens. Due to their colonial habits which implies close contacts between individuals, bats are often parasitized by multiple species of micro-and macroparasites. The particular ecology, behavior, and environment of bat species may shape patterns of intra-and interspecific pathogen transmission, as well as the presence of specific vectorial organisms. This review synthetizes information on a multi-level parasitic system: bats, bat flies and their microparasites. Bat flies (Diptera: Nycteribiidae and Streblidae) are obligate, hematophagous ectoparasites of bats consisting of ~500 described species. Diverse parasitic organisms have been detected in bat flies including bacteria, blood parasites, fungi, and viruses, which suggest their vectorial potential. We discuss the ecological epidemiology of microparasites, their potential physiological effects on both bats and bat flies, and potential research perspectives in the domain of bat pathogens. For simplicity, we use the term microparasite throughout this review, yet it remains unclear whether some bacteria are parasites or symbionts of their bat fly hosts.
Bats are the second most diverse mammalian group after rodents, with ~1390 recognized species across 227 genera (1). Many bat species play keystone roles in ecosystems, where they are essential to pollination, seed dispersal, and pest control (2). Several studies have also highlighted their prominent role as pathogen-reservoirs (3, 4); viruses being the best studied due to their potential as human pathogens (3, 5 – 8). Bats host more viruses per species than rodents, making them an interesting system for both disease ecology and public health research (4, 9).
Bacteria (such as Bartonella spp. and Borrelia spp.) and protozoans (such as Trypanosoma spp. and Plasmodium spp.) have also been detected in bats (8, 10, 11). In recent years, bat-associated Bartonella genotypes have been found in humans, indicating the public health importance of this parasite in bats (12 – 14). Bartonella and other pathogen transmission from bats to humans may occur through religious activities in caves, bat consumption or contact with contaminated products (12, 15). There are documented cases of bat-specific ectoparasites biting humans (16, 17), increasing the potential of bat-born pathogen transmission. Additionally, bat-associated pathogen, such as Trypanosoma cruzi genotype has also been found in humans (18).