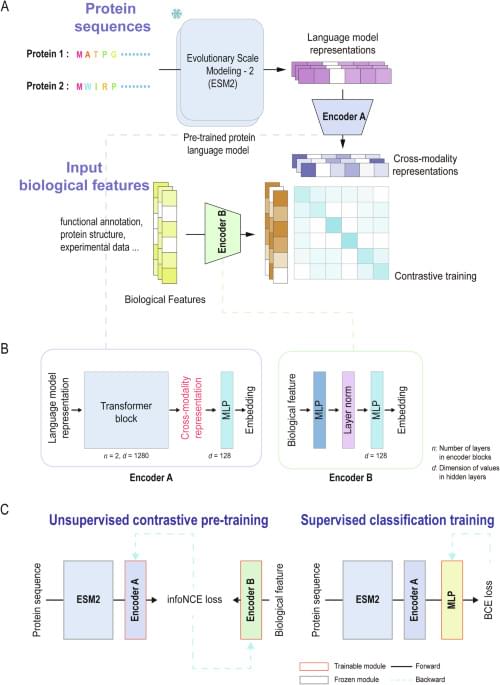
Identifying and characterizing secreted virulence proteins are fundamental for deciphering microbial pathogenicity. Here, the authors introduce a practical training framework to improve protein language model representations by integrating biological features and prior information through contrastive learning.