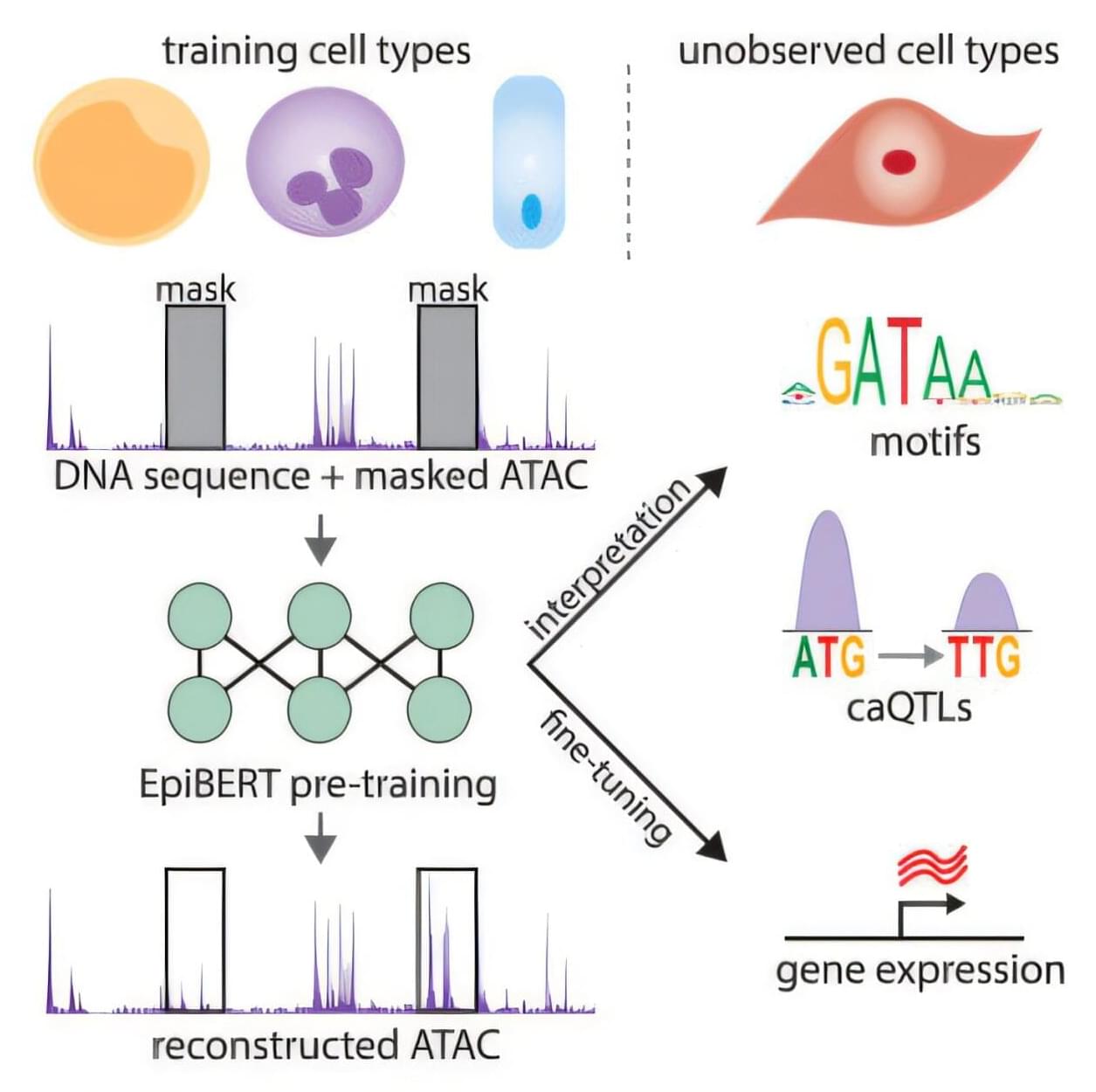
A team of investigators from Dana-Farber Cancer Institute, The Broad Institute of MIT and Harvard, Google, and Columbia University have created an artificial intelligence model that can predict which genes are expressed in any type of human cell. The model, called EpiBERT, was inspired by BERT, a deep learning model designed to understand and generate human-like language.
The work appears in Cell Genomics.
Every cell in the body has the same genome sequence, so the difference between two types of cells is not the genes in the genome, but which genes are turned on, when, and how many. Approximately 20% of the genome codes for regulatory elements determine which genes are turned on, but very little is known about where those codes are in the genome, what their instructions look like, or how mutations affect function in a cell.