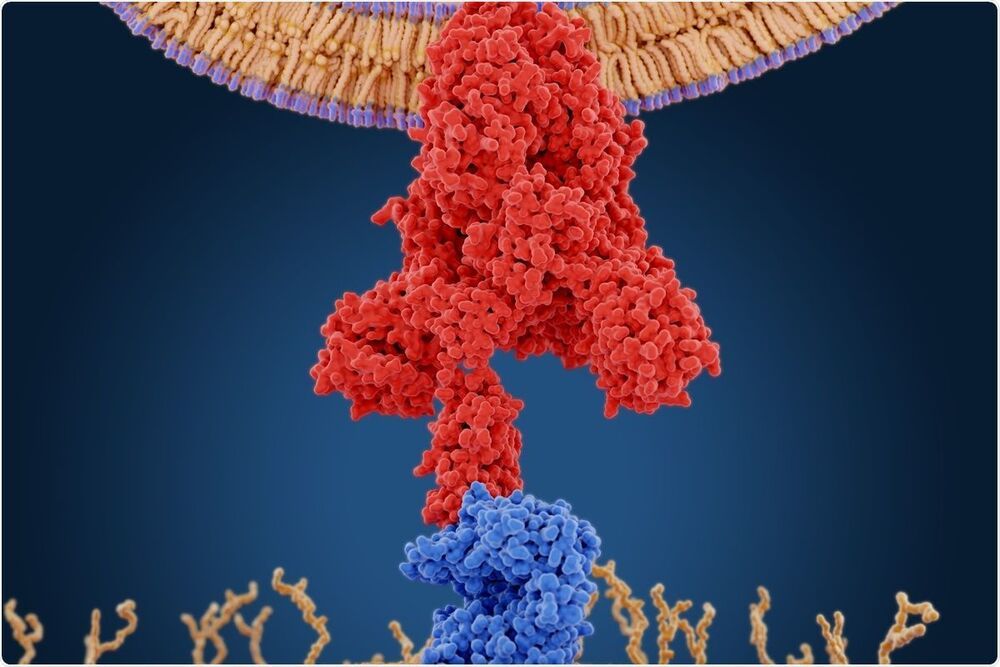
“The furin cleavage site consists of four amino acids PRRA, which are encoded by 12 inserted nucleotides in the S gene. A characteristic feature of this site is an arginine doublet. This insertion could have occurred by random insertion mutation, recombination or by laboratory insertion. The researchers say the possibility of random insertion is too low to explain the origin of this motif. Surprisingly, the CGGCGG codons encoding the two arginines of the doublet in SARS-CoV-2 are not found in any of the furin sites in other viral proteins expressed by a wide range of viruses. Even within the SARS-CoV-2, where arginine is encoded by six codons, only a minority of arginine residues are encoded by the CGG codon. Again, only two of the 42 arginines in the SARS-CoV-2 spike are encoded by this codon — and these are in the PRRA motif. For recombination to occur, there must be a donor, from another furin site and probably from another virus. In the absence of a known virus containing this arginine doublet encoded by the CGGCGG codons, the researchers discount the recombination theory as the mechanism underlying the emergence of PRRA in SARS-CoV-2.”
The ongoing pandemic of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has largely defied attempts to contain its spread by non-pharmaceutical interventions (NPIs). With the massive loss of life and economic damage, the only way out, in the absence of specific antiviral therapeutics, has been the development of vaccines to achieve population immunity.
A new study on the Preprints server discusses the origin of the furin cleavage site on the SARS-CoV-2 spike protein, which is responsible for the virus’s relatively high infectivity compared to relatives in the betacoronavirus subgenus.