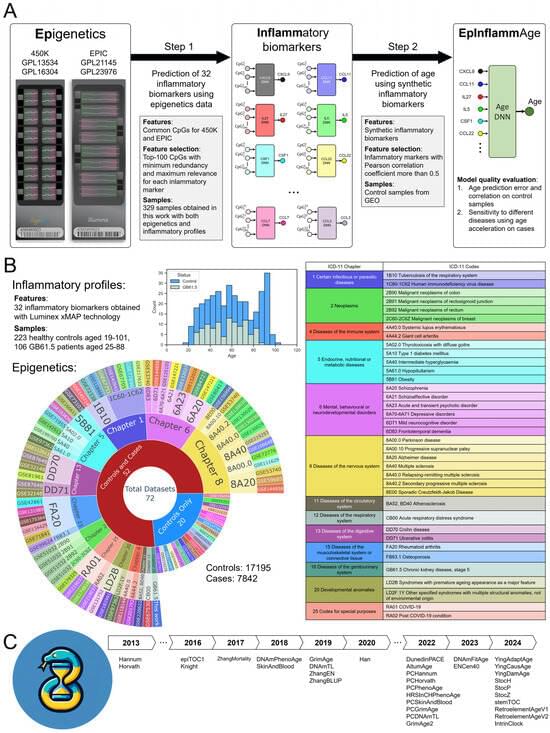
We present EpInflammAge, an explainable deep learning tool that integrates epigenetic and inflammatory markers to create a highly accurate, disease-sensitive biological age predictor. This novel approach bridges two key hallmarks of aging—epigenetic alterations and immunosenescence. First, epigenetic and inflammatory data from the same participants was used for AI models predicting levels of 24 cytokines from blood DNA methylation. Second, open-source epigenetic data (25 thousand samples) was used for generating synthetic inflammatory biomarkers and training an age estimation model. Using state-of-the-art deep neural networks optimized for tabular data analysis, EpInflammAge achieves competitive performance metrics against 34 epigenetic clock models, including an overall mean absolute error of 7 years and a Pearson correlation coefficient of 0.85 in healthy controls, while demonstrating robust sensitivity across multiple disease categories. Explainable AI revealed the contribution of each feature to the age prediction. The sensitivity to multiple diseases due to combining inflammatory and epigenetic profiles is promising for both research and clinical applications. EpInflammAge is released as an easy-to-use web tool that generates the age estimates and levels of inflammatory parameters for methylation data, with the detailed report on the contribution of input variables to the model output for each sample.