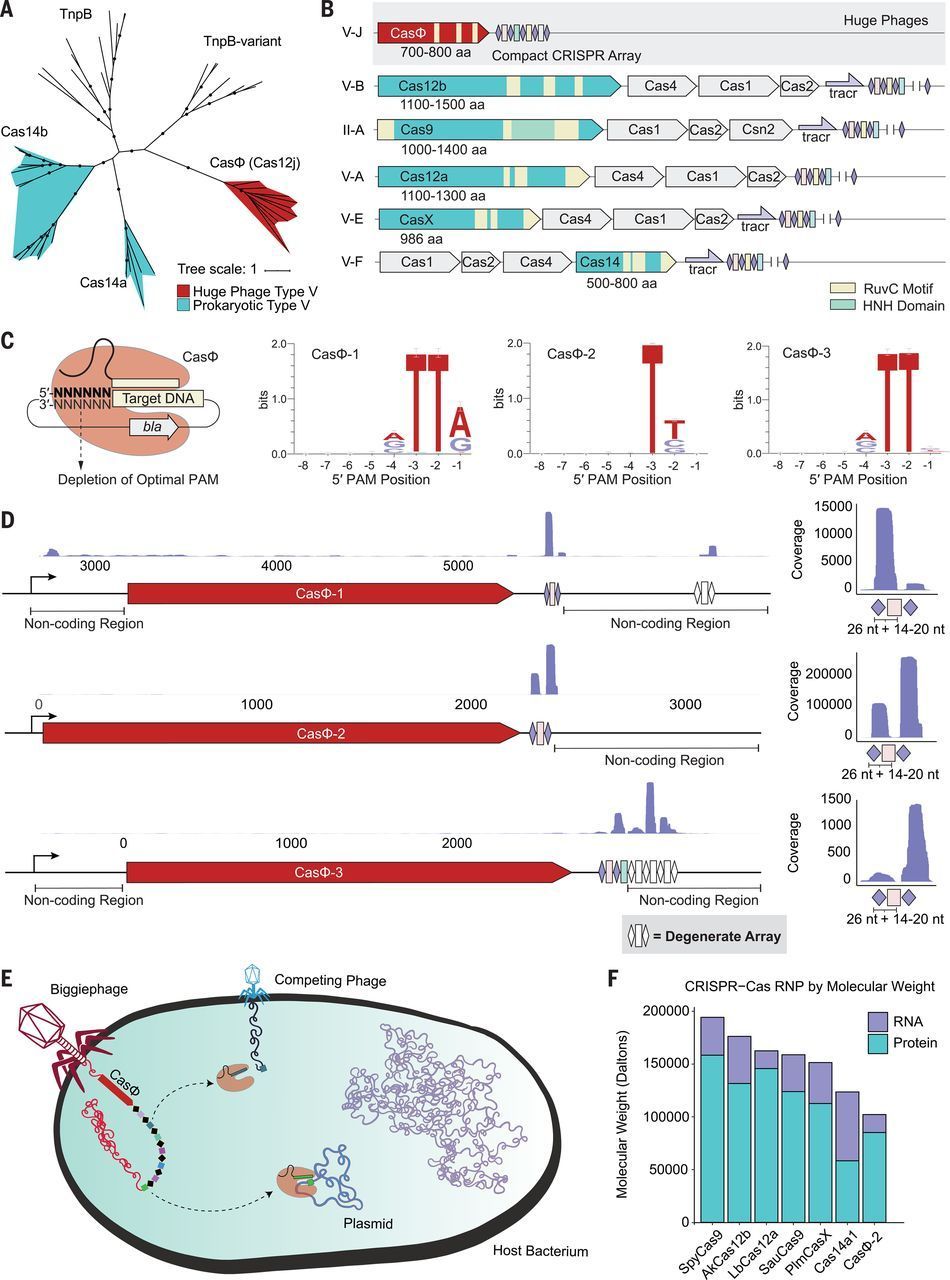
The CRISPR-Cas system, naturally found in many prokaryotes, is widely used for genome editing. CRISPR arrays in the bacterial genome, derived from the genome of invading viruses, are used to generate a CRISPR RNA that guides the Cas enzyme to destroy repeat viral invaders. Recently, an unexpectedly compact CRISPR-Cas system was identified in huge bacteriophages. Pausch et al. show that even though this system lacks commonly found accessory proteins, it is functional. In addition to a CRISPR array, the only component of the system is an enzyme called CasF, which uses the same active site to process transcripts of the CRISPR arrays into CRISPR RNA and to destroy foreign nucleic acids. This system, which is active in human and plant cells, provides a hypercompact addition to the genome-editing toolbox.
Science this issue p. 333
CRISPR-Cas systems are found widely in prokaryotes, where they provide adaptive immunity against virus infection and plasmid transformation. We describe a minimal functional CRISPR-Cas system, comprising a single ~70-kilodalton protein, CasΦ, and a CRISPR array, encoded exclusively in the genomes of huge bacteriophages. CasΦ uses a single active site for both CRISPR RNA (crRNA) processing and crRNA-guided DNA cutting to target foreign nucleic acids. This hypercompact system is active in vitro and in human and plant cells with expanded target recognition capabilities relative to other CRISPR-Cas proteins. Useful for genome editing and DNA detection but with a molecular weight half that of Cas9 and Cas12a genome-editing enzymes, CasΦ offers advantages for cellular delivery that expand the genome editing toolbox.