As the world population is ageing rapidly, with over two billion people projected to be above the age of 60 by 2050, age-related brain disorders are on the rise. Living longer but in poor health is not only a daunting prospect, it also places a substantial burden on healthcare systems worldwide. The idea of being able to counteract the functional decline of our brain through rejuvenating interventions sounds therefore promising. The question is how can we identify compounds that have the potential to efficiently rejuvenate brain cells and to protect the ageing population from neurodegeneration? Prof. Antonio Del Sol and his teams of computational biologists, based both at the LCSB from the University of Luxembourg and at the CIC bioGUNE in Bilbao, used their machine learning expertise to tackle the challenge.
The researchers developed what is called an “ageing clock”, a computational tool designed to measure the biological age of cells, as opposed to their chronological age. Indeed, the organs and tissues of people of the same age can evolve differently over time depending on genetic and environmental factors, leading to different biological ages. These clocks are therefore useful tools to assess ageing at the molecular level and can help in understanding its causes and consequences.
The clock designed by the LCSB and CIC bioGune researchers is specific to the brain and uses gene expression information from 365 genes to make predictions. Using a machine learning approach, it was trained on data from healthy individuals, aged from 20 to 97, and could accurately predict their age. Further tests showed that the clock is able to estimate the biological age of different cell types in the brain, especially neurons. Lastly, by looking at the predicted biological ages for healthy individuals and for patients with neurological conditions, the researchers observed that patients exhibited a higher biological age.
“Our results tell us that the biological age of the brain cells calculated by our clock reflects the decline in brain function experienced by the patients, especially between 60 and 70, and is even correlated with the degree of neurodegeneration,” explains Dr Guillem Santamaria, first author of the study. “It supports the view of neurodegeneration as a form of accelerated ageing but, more importantly, the positive association between neurodegeneration and biological age suggests that the rejuvenating interventions identified by the clock could serve as neuroprotective agents.”
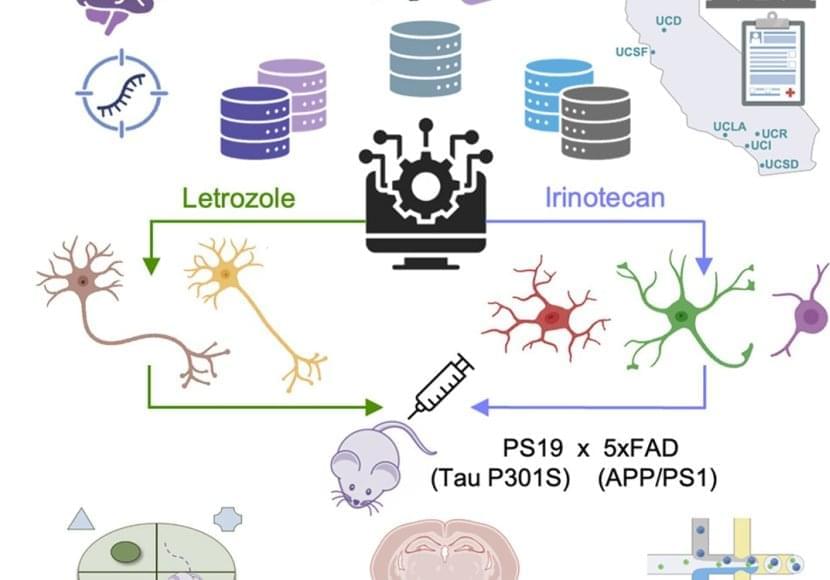
The aim of the researchers was to use the clock to find genetic or chemical interventions that would significantly shift back the biological age of brain cells. They explored the effect of thousands of compounds on neural progenitor cells and neurons and identified 453 unique rejuvenating interventions.
Among the identified compounds that have the potential to reverse the biological age of the two types of brain cells, several are known to extend lifespan in animal models and some are already used to treat neurological disorders, but the vast majority has not yet been studied in the context of health-or lifespan extension. “On the one hand, the fact that our computational platform identified drugs that have a known effect on brain function supports the idea that using the predicted effect of a compound on the biological age is an efficient way to evaluate its neuroprotective potential,” details Prof. Antonio Del Sol, head of the Computational Biology groups at the LCSB and CIC BioGUNE. “On the other, the results also highlight that our clock can help us find many new candidates that haven’t been studied before for their rejuvenating properties. It opens up a lot of new avenues.”
As a proof of concept of their approach, the researchers then tested three of the predicted compounds in mice, in collaboration with the team of Prof. Rubén Nogueiras at the Centre for Research in Molecular Medicine and Chronic Diseases. The administration of these drugs significantly reduced anxiety and slightly increased spatial memory in older mice, addressing two well-known symptoms associated with ageing. An analysis of gene expression showed that the combination of these compounds also led to a shift toward a younger phenotype. Altogether, these results show that a selection of compounds predicted to rejuvenate the brain did produce rejuvenation at the molecular level in the cortex of aged mice and had an impact on behavioural and cognitive functions.
Globally, the study, recently published in the journal Advanced Science, highlights the computational ageing clock developed by the researchers as a valuable resource for identifying brain-rejuvenating interventions with therapeutic potential in neurodegenerative diseases. It provides a strong foundation for further research. “The hundreds of compounds predicted by our platform require validation across multiple biological systems to assess their efficacy and safety, offering extensive opportunities for future therapeutic development,” concludes Prof. Antonio Del Sol.